IJCRR - 9(3), February, 2017
Pages: 53-57
Date of Publication: 10-Feb-2017
Print Article
Download XML Download PDF
Microbial Fingerprinting - A current vogue in Microbial Forensics
Author: Moumita Sinha1, I. Arjun Rao2
Category: General Sciences
Abstract:Classification and Identification of the microorganisms are of utmost importance in the field of environmental, industrial, medical and agricultural microbiology, microbial ecology and microbial forensic studies. Conventional phenotype-based methods come across many challenges and shortcomings which limit their usability. Molecular techniques offer better arrangements in recognizing and portraying microorganisms. A few DNA fingerprinting strategies have been produced and are being used as of now. In principle, most of these methods are based on PCR and restriction site analysis. Some of these methods are still not cost-effective in use and require huge set-up cost. Continuous research is going on around the world to improve the methodology and applicability of these methods as well as to make them economic and in routine use in forensic investigations..
Keywords: Microbes, Fingerprinting, DNA, RFLP, Multilocus
Full Text:
Introduction
Microbiology is the investigation of tiny life forms, or any living life form that is either a solitary cell (unicellular), a cell group, or has no cells by any stretch of the imagination (acellular). Numerous microorganisms (organisms) are pathogenic and cause illness while different microorganisms are not a threat to human wellbeing. Microorganisms can aimlessly and unlawfully be utilized as operators of natural fighting, bio-violations and agro fear based oppression. Worried to this, Microbial Forensics rose as interdisciplinary field of microbiology dedicated to the improvement, assessment, approval, and utilization of strategies to distinguish and completely portray microbial specimens containing a natural specialist or its segments1. The principle objective of criminological microbiologist is to distinguish feasible pathogens and recognize their DNA marks to decide the root of likely source. In the late years, another logical train microbial fingerprinting has been set up under microbial legal sciences with a specific end goal to fortify the law implementation reaction particularly in a bioterrorism occasion 2. Microbial fingerprinting strategies are a class of methods that separate microorganisms or gatherings of microorganisms in light of novel attributes of a general part or area of a bio particle (e.g., phospholipids, DNA, or RNA). Microbial fingerprinting strategies give a general profile of the microbial group, and some can be utilized to distinguish subsets of the microorganisms exhibit. This survey article means to reveal some insight into this new train and the strategies utilized as a part of microbial fingerprinting.
Different Microbial Fingerprinting Methods (MPMs)
Microbial fingerprinting strategies can give a full examination of the microbial group. It doesn't require much learning about which microorganisms are of the premium. The hereditary techniques permit distinguishing proof of primary individuals from the microbial group to the family or even class level. It indicates distinctive microorganisms or gatherings of microorganisms in view of one of a kind attributes of a mutual thing or area of a bio atom, (for example, phospholipids, DNA, or RNA). Microbial Fingerprinting Methods are applicable to give a general perspective of the microbial group. It shows microbial differing qualities and tells about sorts of metabolic procedures happening on the site. It likewise distinguishes a subset of the microorganisms introduces in the specimen. Advance, it can recognize the microbial greenery of the specific topographical area which is particular to that area. This is relevant and important because every microorganism has its own signature flora that can determine the surrounding environment of an individual during the criminal investigation. Some of the methods used for microbial fingerprinting are as follows:
PLFA Analysis
Phospholipids are an essential auxiliary segment of the layers of every living cell and separate quickly upon cell demise. In this manner, the mass of PLFAs in an example is an immediate measure of the feasible biomass in the specimen. While all cell layers contain phospholipids, not all life forms or gatherings of creatures contain the same PLFA sorts in similar extents. A few classes of creatures deliver one of a kind or "mark" sorts of PLFA3. Measuring these PLFA assembles in this manner makes a profile or unique finger impression of the reasonable microbial group and gives knowledge into a few essential microbial utilitarian gatherings (e.g., iron-and sulfate-decreasing microscopic organisms).
PLFA examination is like an evaluation of other concoction mixes present as blends (e.g., unstable natural mixes) in ecological examples: (1) extraction, (2) detachment by gas chromatography with fire ionization discovery, and if important, (3) affirmation of recognizable proof by mass spectroscopy. PLFA investigation can likewise be joined with stable isotope testing (SIP) to show that biodegradation is happening by measuring consolidation of the steady isotope mark into biomass. PLFA examination is economically accessible.
DGGE Analysis
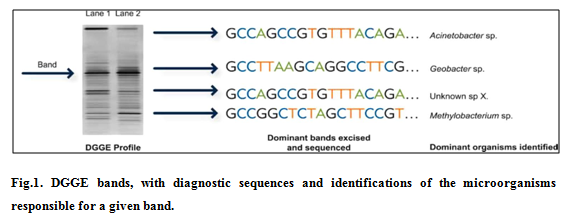
DGGE is a nucleic corrosive (DNA or RNA)–based strategy used to create a hereditary unique mark of the microbial group and possibly distinguish overwhelming microorganisms. DGGE profiles are frequently used to look at contrasts or changes in microbial group differing qualities and structure between tests, after some time or space or in light of treatment. DGGE typically includes a four-stage handle: (1) DNA or RNA extraction, (2) intensification, (3) detachment and representation, and (4) grouping recognizable proof. The enhancement step utilizes polymerase chain response (PCR) to produce a large number of duplicates of a variable locale inside an objective quality. The DNA course of action of this variable region is different for each kind of minute living being. Thus, the PCR step makes a mix of the quality bits every addressing a creature bunches appear in the primary example. The third step of DGGE uses an electric current (electrophoresis) and a denaturing system to separate this mix in light of the DNA gathering, making a profile, or exceptional check, of the microbial gathering. Figure 1 exhibits an ordinary acrylamide gel picture: a subset of the individual "gatherings" are isolated (physically cut) from the gel, the DNA progression is settled for each removed band, and the ensuing DNA plan is stand out from a database to perceive the microbial people identifying with each band 4. Promote understanding is construct to a great extent in light of connecting site conditions and exercises to general attributes of the microorganisms that were recognized in the specimen. DGGE is economically accessible.
T-RFLP Analysis
T-RFLP has also been employed to characterize microbial communities 5. Similar to DGGE, T-RFLP is a nucleic acid (DNA or RNA)–based technique that provides a fingerprint of the microbial community and can be used to identify specific microbial populations. T-RFLP is a four-step process: (1) DNA or RNA extraction, (2) PCR amplification, (3) enzyme digestion, and (4) fragment identification. After isolation of the aggregate group DNA or RNA, PCR amplification with a fluorescent labelled PCR primer is utilized to make different duplicates of an objective quality, and the PCR items are then processed with confinement proteins that cut the DNA particle at known arrangements. The span of each subsequent terminal confinement piece is characteristic of a particular microorganism. T-RFLP offers greater sensitivity than DGGE (i.e., it may detect microorganisms that are present at lower numbers in a sample). T-RFLP is commercially available 6.
Other DNA based Advanced Microbial Fingerprinting Methods
Restriction endonuclease analysis of chromosome (REAC)
REAC includes disconnection of chromosomal DNA, processing with at least one limitation compounds took after by their determination into detectable banding examples or "unique mark" after electrophoresis on agarose or polyacrylamide gel. Many pieces running from 0.5-5 kb long are created. The groups acquired are recolored in situ or denatured inside the gel, smeared onto a reasonable layer (nitrocellulose/nylon) and after that recolored. The subsequent example of groups, mirroring the cutting destinations of the specific catalysts in the chromosome, is exceedingly normal for the given strain and is alluded to as the strain's "unique mark". Strains are separated in light of their fingerprints. Separates indicating one band contrast in the unique mark are considered subtypes of each other. Designs having at least 2 band contrasts are for the most part described as various strains. The varieties in the fingerprints acquired by these techniques represent even the minor changes in the hereditary substance of an organism like point transformation, additions, erasures, site particular recombination and change. In this manner, REAC is an exceedingly delicate method and even a solitary occasion bringing about an adjustment in DNA can be followed, along these lines it is a critical apparatus for epidemiological examination for strain writing. REAC helped in following the flare-up of Pseudomonas aeruginosa mastitis among Irish Dairy groups 7. One noteworthy favorable position of this technique is that it includes the entire chromosome, so the strains are thought about on an expansive premise and no earlier learning of grouping information is required. Utilizing REAC all strains can be written. In any case, at times fingerprints are extremely mind boggling and hard to translate. Nearness of plasmid DNA in the response blend can at times meddle with the outcomes. As of late a few PC based examination strategies have been produced that make correlation of REAC examples simple and develop databases for identity searches and epidemiological typing 6.
Restriction endonuclease analysis of plasmid DNA (REAP)
This strategy can be utilized for those microorganisms which harbor plasmids. Plasmids can be recognized promptly by basic lysis system took after by the agarose gel electrophoresis of the lysate 8. The numbers and the sizes of the plasmids present are utilized as the premise of strain recognizable proof. This strain writing method has been utilized effectively for investigation of episodes of nosocomial diseases and group procured contaminations brought about by different types of gram-negative microbes 9-10. A few strains of microscopic organisms convey just a solitary huge plasmid, in the scope of 100-150kb. Due to the trouble to separate plasmids in this range, a limitation endonuclease processing step is added to expand the unfair force of agarose gel electrophoresis. At present, this technique is fundamentally utilized for staphylococcal secludes, which regularly convey different plasmids and for chose types of Enterobacteriacea, which frequently have extensive unmistakable plasmids 11.
Random amplified polymorphic DNA (RAPD)
This strategy depends on self-assertive intensification of polymorphic DNA groupings. Enhancement is completed utilizing single or different, non-particular preliminaries whose successions are arbitrary and not intended to be reciprocal to a specific site in the chromosome. These preliminaries tie at different 'best-fit' groupings on the denatured DNA under low stringency conditions and stretch out productively to give short amplicons. In consequent cycling, conditions are made more stringent so preparation keeps on binding to best-fit successions and create results of settled lengths. Their (items) electrophoresis and recoloring produces the unique mark. RAPD has the fundamental preferred standpoint that no earlier grouping data is required. Additionally, the whole genomic succession is investigated for correlation. Since the preparation are not coordinated against a specific hereditary locus, a few preparing occasions can come about because of varieties in trial conditions, making thorough institutionalization of the technique basic. The significant detriment of this strategy is absence of between research facility reproducibility. A little change in convention, preparation, polymerase or DNA extraction may give distinctive outcomes.
RAPD has been utilized for writing of various microscopic organisms utilizing 10-mer preparation (oligonucleotides comprising of 10 nucleotides). It was completed for Campylobacter coli and C. jejuni 12, Listeria monocytogenes 13, Staphylococcus haemolyticus 14, Vibrio vulnificus 15. In V. vulnificus writing, prejudicial force of RAPD was highlighted; a distinction in band examples was acquired amongst exemplified and non-embodied isogenic morphotypes. Another adaptation of RAPD is AP-PCR i.e. self-assertively prepared PCR in which PCR is completed with subjective preparation. Here, PCR is completed utilizing >20-mer preparation rather than 10-mer (RAPD). Different subtle elements of the technique stay comparative16-18. As of late, to advance unwavering quality and reproducibility of subjectively prepared PCR, different methods have been suggested by Tyler et al. (1997) 19.
Multilocus sequence typing (MLST)
MLST depends on the direct sequencing of ~500 nucleotides of various housekeeping qualities. The grouping of each of these quality sections is considered as a one of a kind allele, and dendrograms are built from the pair wise distinction in the multilocus allelic profiles by bunch examination. As this technique records the varieties that amass gradually, MLST is a reasonable strategy to concentrate long haul and worldwide the study of disease transmission or development of organisms. Likewise, MLST is appropriate for the development of worldwide databanks assessable to various research facilities for result examination and arrangement 20. As of late MLST has turned into a best quality level method for grouping of Neisseria meningitides 20. MLST has likewise been set up for Streptococcus pneumonia 21-22 and is in advance for Streptococcus pyogenes, Haemophilus influenzae and Campylobacter jejuni. As of late Cocolin et al. (2000) 23 have utilized a comparable method for recognizing 39 strains of Lactobacillus species confined from normally aged Italian frankfurters. A little section from 16S rRNA was increased through PCR. Tannock et al. (1999) 24 distinguished Lactobacillus secludes from gastrointestinal tract, silage and yogurt by opening up and sequencing spacer district in the vicinity of 23S rRNA qualities (The successions got were contrasted and the reference strains in databases, for example, Genbank and a likeness of at least 97.5% was considered to give ID). The 16S-23S intergenic spacer area sequencing has been utilized for recognizable proof of Clostridium difficile and Staphylococcus aureus 25. Albeit costly and work concentrated, none of the DNA fingerprinting procedure depicted above is as dependable or as reproducible as MLST. Once the cost and the trouble of the huge scale sequencing have been lessened by innovative improvements, MLST will turn into the technique for decision in numerous research centers far and wide.
Low-Molecular-Weight (LMW) RNA fingerprinting
A hereditary fingerprinting strategy that has been utilized for over 10 years is profiling of low-atomic weight (LMW) RNA (5S ribosomal RNA {rRNA}and exchange RNA {tRNA} 26. The method 26 is clear; add up to RNA is extricated from a natural specimen, and isolated by high-determination polyacrylamide gel electrophoresis. The division profiles of the 5S rRNA and tRNA (the 16S r RNA particles are too large to enter the gel) can be pictured by silver recoloring or via autoradiography if the RNA was radioactively named. Accordingly, the profiles are checked, and put away in an electronic database for correlation. LMW RNA profiling has been utilized to screen bacterial populace elements in an arrangement of freshwater mesocosms after expansion of non-indigenous microorganisms and culture medium 27. The approach was likewise used to explore the assorted qualities and movement of bacterial populaces in a stratified water segment of the focal Baltic Sea 28. (Höfle and Brettar, 1996). Bidle and Fletcher (1995) 29 utilized LMW RNA profiling to think about free-living and molecule related bacterial groups from various profundities and distinctive destinations in an estuary straight. Favorable position of the LMW RNA fingerprinting method is the nonappearance of an in vitro enhancement venture to deliver adequate material to be broke down, in light of the fact that such an intensification step may make mistakes 30. Another positive point is that individual groups can be sequenced 31 or profiles can be hybridized with particular tests to evaluate the personality of the group individuals. In any case, just constrained phylogenetic data can be acquired from the little 5S rRNA (max. 131 nucleotides) and tRNA (max. 96 nucleotides). Another frail point is the quick debasement of RNA, which may shape extra groups in the profiles making the translation of results troublesome.
Importance of Data Generated from Microbial Fingerprinting Methods
Information produced from MPMs is utilized to comprehend which microorganisms are available and how they are associated with their natural conditions. MPMs can likewise be utilized to track the general changes in the microbial group after some time or in light of remediation exercises. The subsequent fingerprints can be coupled to factual examination and different sorts of estimations. Tests after investigation are contrasted with reference groups, which are nearer to reference band are deciphered as being biologically comparable while that are far separated are translated as containing huge contrasts.
Conclusion
From the previous record, plainly in the most recent decade or something like that, few DNA-based fingerprinting strategies have been produced to help with the distinguishing proof and portrayal of the organisms. Despite the fact that the traditional phenotypic strategies like serotyping would keep on being utilized for quite a while to come, atomic procedures will be progressively utilized as a part without bounds. Additionally look into on the techniques of DNA fingerprinting strategies would uncover the pitfalls to which these are inclined and would absolutely be refined, making them more powerful and pertinent in the greater part of the world where research facilities are malpractice. A few of these techniques will then empower production of huge reference libraries or databases of the typed organisms for correlation, fast recognizable proof, portrayal and characterization of new disconnects over the world.
References:
1. Bhatia Mohit, Mishra Bibhabati, Thakur Archana, Dogra Vinita, Loomba Poonam Sood. 2016. Concept of Forensic Microbiology and its Applications. Sikkim Manipal University Medical Journal. Vol. 3(1).
2. Budowle. B, Schutzer SE, Einseln A, Kelley LC, Walsh AC, Smith JA, et al. 2003. Building microbial forensics as a response to bioterrorism. Science. Public health. 301 (5641), 1852-3.
3. Hedrick David B, Peacock Aaron, Stephen John R, Macnaughton Sarah J, Bruggemann Julia, White David C. Measuring soil microbial community diversity using polar lipid q fatty acid and denaturing gradient gel electrophoresis data. Journal of Microbiological Methods 41 (2000) 235–248.
4. Muyzer G, de Waal EC, Uitterlinden AG. 1993. Profiling of microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59, 695–700.
5. Osborn AM, Moore ERB, Timmis K. 2000. An evaluation of terminal-restriction fragment length polymorphism (T-RFLP) analysis for the study of microbial community structure and dynamics. Environ. Microbiol. 2:39-50.
6. Interstate Toxicology, Regulatory Council, 2011. http:// www.itrcweb.org. Retrived on 17 December, 2016.
7. Daly M, Power E, Bjorkroth J, et al. 1999. Molecular analysis of Pseudomonas aeruginosa epidemiological investigation of mastitis outbreaks in Irish dairy herds. Applied and Environmental Microbiology. 65, 2723 -2729.
8. Tenover FC. 1985. Plasmid fingerprinting a tool for bacterial strain identification and surveillance of nosocomial and community acquired infections. Clinics in Laboratory Medicine. 5, 413-436.
9. Schaberg DR, Tompkins LS, Falkow S. 1981. Use of agarose gel electrophoresis of plasmid deoxyribonucleic acid to fingerprint gram-negative bacilli. Journal of Clinical Microbiology. 13, 1105-1110.
10. Fornasini M, Reeves RR, Murray BE, et al. 1992. Trimethoprim resistant Escherichia coli in households of children attending day care centers. Journal of Infectious Disease. 166, 326-330.
11. Pfaller M A, Wakefield DS, Hollis R, et al. 1991. The clinical microbiology laboratory as an aid in infection control. The application of molecular techniques in epidemiologic studies of methicillin resistant Staphylococcus aureus. Diagnostic Microbiology & Infectious Disease. 14, 209-214.
12. Maidenn MCJ, Bygrives JA, Feil E, et al. 1998. Muitilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proceedings of the National Academy of Sciences. USA. 95, 3140-3145.
13. Czajka J, Bsat N, Piana M, et al. 1993. Differentiation of Listeria monocytogenes and Listeria innocua by 16S genes and intraspecies discrimination of Listeria monocytogenes strains by random amplified polymorphic DNA polymorphisms. Applied and Environmental Microbiology. 59, 304-308.
14. Young KA, Power EG, Dryden MS, et al. 1994. RAPD typing of clinical isolates of Staphylococcus haemolyticus. Letters in Applied Microbiology. 18, 86-89.
15. Warner JM, Oliver JD. 1999. Randomly amplified polymorphic DNA analysis of clinical and environmental isolates of Vibrio vulnificus and other Vibrio species. Applied and Environmental Microbiology. 65, 1141-1144.
16. Welsh J, McClelland M. 1990. Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acid Research. 18, 7213-7218.
17. Welsh J, McClelland M. 1993. The characterization of pathogenic microorganisms by genomic fingerprinting using arbitrarily primed polymerase chain reaction (AP-PCR). In Diagnostic Molecular Microbiology (eds. Persing D. H. et al.) washington: ASM press. pp. 595- 602.
18. Williams JGK, Kubelik AR, Lival KJ, et al. 1990. DNApolymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acid Research. 18, 6531-6535.
19. Tyler KD, Wang G, Tyler SD, Johnson WM. Factors affecting reliability and reproducibility of amplification-based DNA fingerprinting of representative bacterial pathogens. J. Clin. Microbiol., 35 (1997), pp. 339–346.
20. Madden RH, Moran L, Scates P. 1996. Sub-typing of animal and human Campylobacter spp. using RAPD. Letters in Applied Microbiology. 23, 167-170.
21. Enright MC, Spratt BG. 1998. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology. 144, 3049-3060.
22. Enright MC, , Griffiths D, Spratt BG. 1999. The three major Spanish clones penicillin-resistant Streptococcus pneumoniae are the most common clones recovered recent cases of meningitis in Spain. Journal of Clinical Microbiology. 37, 3210-3216.
23. Cocolin L, Manzano M, Cantoni C, Comi G. 2000. Development of a rapid method for the identification of Lactobacillus spp. isolated from naturally fermented Italian sausages using a polymerase chain reactiontemperature gradient gel electrophoresis. Letters in Applied Microbiology. 30, 126-129.
24. Tannock GW, Timisjarvi AT, Rodtong S. 1999. Identification of Lactobacillus isolates from the gastrointestinal tract, silage and yoghurt by 16S-23S rRNA gene intergenic spacer region sequence comparisons. Applied and Environmental Microbiology. 65(9), 4264-4267.
25. Gurtler V, Stanisich, VA. 1996. New approaches to typing and identification of bacteria using the 16S-23S rDNA spacer region. Microbiology, 142, 3-16.
26. Höfle M. 1988. Identification of bacteria by low molecular weight RNA profiles: a new chemotaxonomic approach. J Microbiol Methods 8: 235-248.
27. Höfle MG. 1998. Genotyping of bacterial isolates from the environment using low molecular weight RNA fingerprints. Mol Microb Ecol Manual 3.3.7: 1-23.
28. Höfle M. 1992. Bacterioplankton community structure and dynamics after large-scale release of non-indigenous bacteria as revealed by low molecular weight RNA analysis. Appl Environ Microbiol 58: 3387-3394.
29. Bidle KD, Fletcher M .1995. Comparison of free-living and particle-associated bacterial communities in the Chesapeake Bay by stable low-molecular-weight RNA analysis. Appl Environ. Microbiol 62: 944-952.
30. Wintzingerode F, Göbel UB, Stackebrandt E .1997. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol Rev 21: 213-229.
31. Höfle MG, Brettar I . 1996. Genotyping of heterotrophic bacteria from the central Baltic Sea by use of low-molecular-weight RNA profiles. Appl Environ Microbiol 62: 1383- 1390.
|






 This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License