IJCRR - 13(17), September, 2021
Pages: 131-135
Date of Publication: 12-Sep-2021
Print Article
Download XML Download PDF
Ethidium Bromide-Agar Cartwheel Method in the Detection of Efflux Pump Mediated Multi-Drug Resistance in Enterobacteriaceae
Author: Patil R, Rangappa KG, Rangaiah A, Shankar SM
Category: Healthcare
Abstract:Introduction: Multidrug-resistant (MDR) strains of bacteria pose a major threat in clinical settings. Multidrug resistance can be due to various mechanisms but is primarily the result of over-expressed efflux pumps that extrude unrelated antibiotics before they reach the intended targets. The emergence of MDR due to efflux pumps has to lead to a diagnostic and therapeutic dilemma. Ethidium Bromide (EtBr)-agar cartwheel assay is a newly discovered simple, safe and cost-effective method to determine efflux pump activity. Objectives: The study aimed at the detection of efflux pump activity in MDR strains belonging to Enterobacteriaceae family using the EtBr cartwheel method as well as determining the antimicrobial susceptibility pattern of MDR Enterobacteriaceae. Materials and methods: A total of 95 MDR Enterobacteriaceae isolates from various clinical samples were included in the study. Identification and antimicrobial susceptibility were done following Clinical Laboratory Standards Institute(CLSI) 2019 guidelines. They were evaluated for efflux activity using the EtBr agar cartwheel method. Data analysis was performed using SPSS version 20. Results: All the 95 isolates were tested for efflux pump using the Ethidium Bromide agar cartwheel technique. The results showed that 47(49.47%) of the total isolates gave positive results. Among the 47 efflux pump, positive organisms majority were Klebsiellapneumoniae, followed by Escherichia coli and Proteus mirabilis. Conclusion: Laboratory detection of efflux pumps in bacteria can be effectively done using EtBr agar cartwheel assay. We conclude that over-expression of efflux pumps has led to an alarming rise in drug resistance and necessary steps should be taken to control this problem.
Keywords: Ethidium Bromide, Cartwheel, Efflux Pump, Multidrug resistance, Enterobacteriaceae, Klebsiellapneumoniae
Full Text:
INTRODUCTION:
Bacterial infections have again become a threat due to the rapid emergence of resistant bacteria caused by the overuse and misuse of antibiotics, thus endangering their efficacy. Bacteria resist the action of antibiotics through several mechanisms.1 Out of these, bacterial efflux pumps are becoming a major concern because they provide bacteria with the ability to drive away many structurally unrelated antibiotics, even before their effect begins to onset.2 These pumps are classified into five: ATP binding cassette superfamily (ABC), major facilitator superfamily (MFS), resistance nodulation cell division superfamily (RND), small multidrug resistance family (SMR), multi-antimicrobial extrusion protein family (MATE).3,4,5
The intestinal tracts of humans and animals form the natural habitat for Enterobacteriaceae which are a heterogeneous group of Gram-negative rods. Enterobacteriaceae cause a variety of human infections that can be broadly classified as either enteric diseases or extra-intestinal infections such as urinary tract infections, bacteraemia, and meningitis. Enterobacteriaceae acquired in the hospital environment are often resistant to many antimicrobial agents.6
The evaluation of efflux systems by conventional methods such as the retention of the fluorescent dye ethidium bromide (EtBr) or radio-labelled antibiotics requires specialised instrumentation not usually available in a clinical laboratory. Hence there is a need to develop a fast and cost-effective method for detecting efflux pumps in efflux mediated multidrug-resistant (MDR) bacteria. The EtBr agar cartwheel assay is a newly discovered simple, instrument-free, safe and cost-effective method utilised for the demonstration of efflux pump activity in bacteria. It employs EtBr as the pump substrate that allows the verification of the existence of an over-expressed efflux system. Thus, there is a maximum concentration of EtBr which is effectively extruded by the cells and higher EtBr concentrations will be retained; hence when the bacterial mass is exposed to ultraviolet (UV) light, fluorescence will be detected. The concentration of EtBr that is required to produce fluorescence in bacterial strains over-expressing efflux systems is considerably higher than that which produces fluorescence of the reference strain.5,7
The present study aimed to detect efflux pumps in MDR bacteria belonging to the Enterobacteriaceae family with the help of the EtBr agar Cartwheel assay in a tertiary care centre.
MATERIALS AND METHODS:
This cross-sectional observational study was carried out in the Department of Microbiology, Bangalore Medical College & Research Institute (BMCRI), Bengaluru from samples sourced from the attached hospitals. The sample size was taken as 95 as calculated with d(absolute precision)=8 and p(prevalence)=83%, using the formula: N=4pq/d2.8MDR Enterobacteriaceae isolates from clinical samples sent for culture and antimicrobial susceptibility testing were included in the study. SusceptibleEnterobacteriaceae, Gram-negative organisms other than Enterobacteriaceae and Gram-positive cocci were excluded from the study. Institutional ethical clearance was obtained. Demographic details like name, age, gender, place and other relevant medical details were obtained from request forms sent to the microbiology laboratory and from the medical records department.
Identification and anti-microbial susceptibility were done using VITEK 2 system as per Clinical Laboratory Standards Institute (CLSI) 2019 guidelines.9 MDR isolates were further tested for efflux activity using the Ethidium Bromide Agar Cartwheel method. Escherichia coli (E.coli) ATCC25922 was used as control.
EtBr-agar cartwheel method:
Bacterial strains were grown in 5 mL of appropriate liquid broth until they reached an optical density (OD) 0.6 at 600 nm. The OD of the cultures was adjusted with PBS to 0.5 McFarland standard. Tryptic soy agar plates containing EtBr concentrations ranging from 0 to 2.5 mg/L were prepared on the same day of the experiment and protected from light. The plates were then divided into as many as 12 sectors by radial lines (cartwheel pattern) as exemplified in Flowchart 1. OD adjusted cultures were inoculated on EtBr-agar plates starting from the centre of the plate and spreading towards the edges, as indicated by the arrowheads shown in Flowchart 1. Each plate included at least one reference strain that served as a comparative control. The number of reference strains to be included may be increased to two or more, depending on a given experiment. The swabbed EtBr-agar plates were then incubated at 37°C for 16 hours and examined under a suitable source of UV light, such as a hand-held UV lamp or a UV transilluminator. The minimum concentration of EtBr (MCEtBr) that produced fluorescence of the bacterial mass was recorded(Fig 1).5,7The absence of fluorescence determines the presence of active efflux pumps in MDR strains.
Statistical analysis: Data is entered into an excel spreadsheet and analysed using SPSS version 20. Data are presented as descriptive statistics and analysed using suitable parametric and non-parametric tests. Data is presented in the form of figures, tables, graphs or pie charts.
RESULTS:
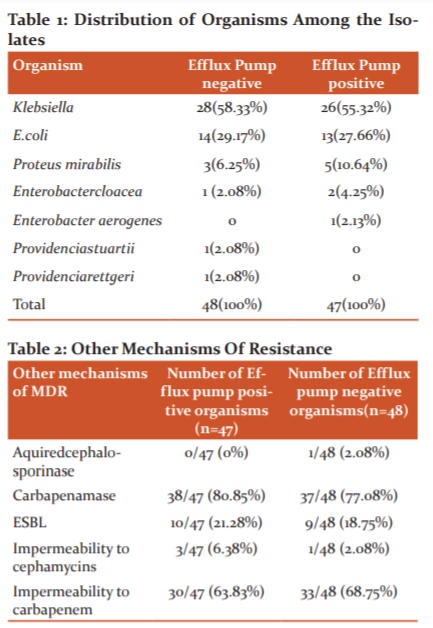
A total of 95 Enterobacteriaceae organisms were isolated from various specimens of patients, of which 54(56.8%) were Klebsiellapneumoniae (K.pneumoniae), 27(28.4%) were E.coli, 8(8.4%) were Proteus mirabilis (P.mirabilis), 3(3.2%) were Enterobacter cloacae, 1(1.1%) was Enterobacteraerogenes, 1(1.1%) was Providenciastuartii and 1(1.1%) was Providenciarettgeri. (Table 1)
83 isolates were recovered from inpatients and 12 isolates were from the outpatient department. The distribution of the patients included in the study among various departments was as follows: 26(27.37%) in Surgical units, 15(15.79%) in Burns ward, 9(9.47%) in Neonatal Intensive Care Unit (ICU), 7(7.37%) in Paediatric units, 6(6.32%) in Obstetrics & Gynaecology (OBG) units, 5(5.26%) in Medicine units, 3(3.16%) in Dermatology units, 2(2.1%) each in Medical ICU, Burns ICU, Pulmonary Medicine units and Paediatric ICU, 1(1.05%) each in Psychiatry, Otorhinolaryngology, OBG ICU, Urology and Orthopaedic units. 10isolates (10.57%) were not traceable.
Specimens included in the study were: pus 48 (50.5%), urine 18 (18.9%), endotracheal aspirate 13(13.7%), Blood 5(5.3%), Sputum 5(5.3%), CSF 2(2.1%), High vaginal/cervical swab 2(2.1%) and fluid aspirates2 (2.1%).
Antimicrobial susceptibility pattern:
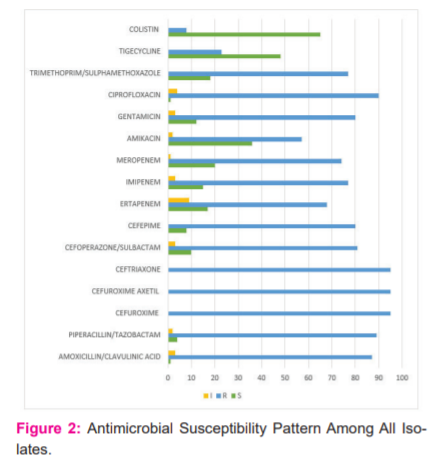
Antibiotic susceptibility to various antimicrobials was done by automated identification/ Antimicrobial susceptibility (ID/AST) method using VITEK-2 (Biomerieux). All 95 (100%) isolates were resistant to cefuroxime, cefuroxime axetil and ceftriaxone. 90 isolates (94.7%) were resistant to ciprofloxacin, followed by resistance to piperacillin/tazobactam (93.68%), amoxicillin/clavulanic acid (91.58%) and cefoperazone/sulbactam (85.26%). Least resistance was noted among the isolates to nitrofurantoin (6.3%) and colistin(8.4%). (Fig 2)
The mean age of patients was 33.21 ± 24.216, M: F ratio was 12:7.
E.coli(27) isolates were most resistant to 2nd and 3rd generation cephalosporins(100%) followed by 4th generation cephalosporins and fluoroquinolones(96.23%) followed by ampicillin (92.56%) with the least resistance to tigecycline and nitrofurantoin(0%).
K.pneumoniae(54) isolates were also most resistant to 2nd and 3rd generation cephalosporins and amoxicillin/clavulanic acid (100%) followed by piperacillin/tazobactam(98.15%) and cefoperazone/sulbactam (94.44%). Colistin resistance was lower among these isolates (3.70%).
P.mirabilis (8) isolates were most resistant to 2nd and 3rd generation cephalosporins, amoxicillin/clavulanic acid, imipenem, amikacin, gentamicin and ciprofloxacin(100%) followed by ampicillin (87.5%) and trimethoprim/sulphamethoxazole (75%). They were least resistant to nitrofurantoin (0%) and tigecycline (12.5%).
The organisms were also flagged for various mechanisms of resistance by VITEK-2. 75(78.94%) isolates were flagged for carbapenemase. 63 (66.31%) showed impermeability to carbapenems. 19(20%) were ESBL producers, 4(4.21%) showed impermeability to cephamycins and 1(1.05%) isolate showed acquired cephalosporins activity. (Table 2)
Efflux pump activity:
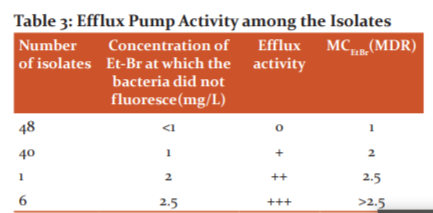
All the 95 isolates were tested for efflux pump using the Ethidium Bromide agar cartwheel technique. The results showed that 47(49.47%) isolates gave positive results out of which 6 did not show fluorescence even at 2.5mg/L(max concentration of EtBr used), 1 isolate did not show fluorescence at 2mg/L but fluoresced at 2.5mg/L and 40 did not fluoresce at 1mg/L only but fluoresced at 2 and 2.5mg/L. (Table 3)
The mean age of the patients who had infections due to efflux pump positive organisms was 34.02± 25.010 and the male: female (M: F) ratio was 30:17.
Among the 47 efflux pump positive organisms 26(55.32%) were K.pneumoniae, 13(27.66%) were E.coli, 5(10.67%) were P.mirabilis, 2(4.25%) were Enterobactercloaceaand 1(2.13%) were Enterobacteraerogenes (Table 1). The efflux pump positive isolates were mostly from pus (56.45%), followed by urine (19.15%).
13(27.7%) of the efflux pump positive isolates were from surgical cases and 7 (14.9%) were from the burns ward. 38(80.85%) isolates were recovered from inpatients and 9(19.15%) isolates were from the outpatient department. 9 (19.15%) isolates were from ICU.
Among the efflux pump positive organisms, some of the organisms were flagged for other mechanisms of resistance like carbapenemase, impermeability to carbapenems, ESBL and impermeability to cephamycins, by Vitek-2. (Table 2)
The most commonly used antimicrobials among the patients whose specimens yielded efflux pump positive isolates were ceftriaxone, piperacillin/tazobactam and amikacin.
DISCUSSION:
Management of infections is very challenging due to the emergence of multidrug resistance. There are very few available options among anti-microbial agents against MDR Gram-negative bacteria, thus posing a major public health threat. Since MDR is primarily the result of over-expressed efflux pumps that extrude unrelated antibiotics before they reach the intended targets, clinical laboratories should develop and implement new and improved methods for the timely identification of efflux mediated MDR phenotypes.
95 organisms were tested for efflux pump activity, which is very high as compared to studies conducted by Rana T et al. and Al Fayyadh Z et al. and Martins M et al..5,7,10
-
Efflux Pump Activity:-
In the present study, EtBr fluorescence was not observed in 47/95 (49.47%) MDR isolates, which suggested that these isolates contain efflux pump which effluxed out EtBr from the bacterial cell. 6 isolates showed over-expressed efflux systems by not showing fluorescence even at 2.5mg/L(highest concentration of EtBr used), 1 isolate showed intermediate efflux activity by showing fluorescence at 2.5mg/L but not at 2mg/L and 1mg/L while 40 showed mild efflux pump activity by showing fluorescence at 2 and 2.5mg/L.
In a study conducted by Martins M et al., 42 clinical isolates with a confirmed MDR phenotype were evaluated for efflux pump activity by the EtBr cartwheel method. The study included 10 Escherichia coli, 18 Enterobacteraerogenes, 10 Staphylococcus aureus, and 4 Enterococcus faecalis strains. The study findings revealed a presence of efflux activity in 36% of the isolates, which is comparable to the results of our study. Among the efflux pump positive organisms, maximum efflux activity was shown by E.coli(16.6%) species, whereas in our study K.pneumoniae exhibited maximum efflux activity (27.66%).7
In a study conducted by Al Fayyadh Z et al.10 which included 165 specimens from different sources, 93 isolates were identified as E.coli. About 40E.coli isolates were tested for the presence of efflux pump using the cartwheel method. 31 isolates (77.5%) in this study revealed positive results. In our study, the efflux pump activity was relatively lower (49.47%), the reason for which could be the presence of other mechanisms of multidrug resistance.
All 47 efflux pump isolates in the present study were noted to possess one or more of the other mechanisms of resistance: carbapenemase, ESBL, impermeability to cephamycins and impermeability to carbapenem, as revealed by Vitek-2. The other 48 isolates which did not show efflux pump activity also flagged for various other mechanisms of resistance mentioned above along with acquired cephalosporins.
A study by Suwantarat N et al.11shows that one of the major contributors to anti-microbial resistant bacteria in south-east Asia is MDRGram negative bacteria. Overuse of carbapenem therapy to treat these infections has led to the high prevalence of ESBLs in this region. To control the spread of MDRGNs in this region, it is pertinent to improve the infection control practices, have better laboratory detection facilities and advocate judicious use of anti-microbial agents.
-
Anti-microbial susceptibility pattern among Enterobacteriaceae:-
In our report, K.pneumoniae(28.4%) is the most common etiological agent of MDR infections followed by E.coli(8.4%). These organisms were most commonly isolated from pus (50.5%) and urine (18.9%) samples. The majority of them were obtained from Surgery (27.37%) and Burns department(15.79%).
But in a study conducted by Beyene D et al.12 in which 94.5% of the isolates were MDR, E.coliwas the most common etiological agent followed by K.pneumoniae. The majority of the isolates were from urine (62.5%) and by blood (28.4%) and 73% were from ICU.
The mean age of patients in our study was 33.21 ± 24.216 years and the male: female ratio was 12:7. But in a study conducted by M.A. Rajiet al.13, the mean age was 42.4 years with an insignificant difference between the isolates collected from males and females.
These differences could be due to variations in geographic areas, periods of study, target population and sample size.
The isolates encountered in the present study were most resistant to second and third-generation cephalosporins, fluoroquinolones, piperacillin/tazobactam and least resistant to colistin, tigecycline and amikacin (Fig 2). These results are consistent with the results of a study conducted by Charan et al.14where the organisms were most resistant to amoxicillin/clavulanic acid, 2ndand 3rdcephalosporins and carbapenems.
The antimicrobial susceptibility findings related toE.coli and K.pneumoniae are consistent with a study conducted by Lai CC et al.15
CONCLUSIONS:
Laboratory detection of efflux pumps in bacteria can be effectively done using EtBr cartwheel assay. This is a simple and instrument-free technique that can be performed in most laboratories. Based on the present study, it can be concluded that overexpression of efflux pumps has led to an alarming rise in multi-drug resistance and necessary steps should be taken to control this problem. This is expected to aid in controlling hospital-acquired infections and advocating rational use of antimicrobials.
ACKNOWLEDGEMENTS:
The authors express a deep sense of gratitude to faculty and residents, the Department of Microbiology, BMCRI for their guidance and technical assistance during the study. We also acknowledge the support from clinical departments and patients whose clinical specimens were included in the study.
CONFLICT OF INTEREST: Nil
SOURCE OF FUNDING: Research grant received from Rajiv Gandhi University of Health Sciences(RGUHS), Karnataka under the UG Short Term Research 2019 program.
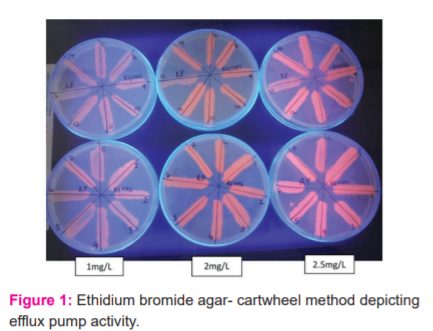
Note: Isolate 13 shows no fluorosence at concentration of 1mg/L and 2mg/L and minimal fluorosence at 2.5mg/L as compared to other isolates. This indicates that isolate 13 contains efflux pumps.
References:
-
Ventola CL. The antibiotic resistance crisis: part 1: causes and threats. P T. 2015;40(4):277-83.
-
Li XZ, Nikaido H. Efflux-mediated drug resistance in bacteria: an update. Drugs. 2009;69(12):1555-1623.
-
Poole K. Efflux pumps as antimicrobial resistance mechanisms. Ann Med. 2007;39(3):162-176.
-
Blair JM, Richmond GE, Piddock LJ. Multidrug efflux pumps in Gram-negative bacteria and their role in antibiotic resistance. Future Microbiol. 2014;9(10):1165-1177.
-
Rana T, Kaur N, Farooq U, Khan A, Singh S. Efflux as an arising cause of drug resistance in Punjab(India). Int J Biol, Pharm, Allied Sci. 2015;4(9): 5967-5979.
-
Jawetz E, Melnick JL, Adelberg EA, Carroll KC. Jawetz, Melnick and Adelberg’s Medical Microbiology. 27thed.New York, NY: McGraw-Hill Education;2016;231
-
Martins M, Viveiros M, Couto I. Identification of efflux pump-mediated multidrug-resistant bacteria by the ethidium bromide-agar cartwheel method. In Vivo. 2011;25(2):171-178.
-
Helmy OM, Kashef MT. Different phenotypic and molecular mechanisms associated with multidrug resistance in Gram-negative clinical isolates from Egypt. Infect Drug Resist. 2017;10:479-498. Published 2017 Dec 8.
-
CLSI. Performance Standards for Antimicrobial Susceptibility Testing. 29th edition.CLSI supplement M100. Wayne, PA: Clinical and Laboratory Standards Institute;2019.
-
Al-Fayyadh ZH, Turkie AM, Al-Mathkhury HJF. New mutations in GyrAgene of Escherichiacoli isolated from Iraqi patients. Iraqi J Sci.2017;58(2B):778-788.
-
Suwantarat N, Carroll KC. Epidemiology and molecular characterization of multidrug-resistant Gram-negative bacteria in Southeast Asia. Antimicrob Resist Infect Control. 2016;5:15. Published 2016 May 4.
-
Beyene D, Bitew A, Fantew S, Mihret A, Evans M. Multidrug-resistant profile and prevalence of extended-spectrum β-lactamase and carbapenemase production in fermentative Gram-negative bacilli recovered from patients and specimens referred to National Reference Laboratory, Addis Ababa, Ethiopia. PLoS One. 2019;14(9):e0222911. Published 2019 Sep 25.
-
Raji MA, Jamal W, Ojemhen O, Rotimi VO. Point-surveillance of antibiotic resistance in Enterobacteriaceae isolates from patients in a Lagos Teaching Hospital, Nigeria. J Infect Public Health. 2013;6(6):431-437.
-
Mulla S, Charan J, Panvala T. Antibiotic sensitivity of Enterobacteriaceae at a tertiary care centre in India. Chron Young Sci. 2011;2(4):214-218.
-
Lai CC, Chen YS, Lee NY. Susceptibility rates of clinically important bacteria collected from intensive care units against colistin, carbapenems, and other comparative agents: results from Surveillance of Multicenter Antimicrobial Resistance in Taiwan (SMART). Infect Drug Resist. 2019;12:627-640. Published 2019 Mar 14.
|






 This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License