IJCRR - 4(1), January, 2012
Pages: 44-50
Print Article
Download XML Download PDF
ISOLATION OF SALMONELLA TYPHI FROM DIFFERENT WATER SOURCES AND IDENTIFICATION
OF DRUG RESISTANT GENES CAT P AND TEM.
Author: Sita Lakshmi T, Geetha R.V, Devi.G, Anitha Roy
Category: Healthcare
Abstract:The aim of the present study was to isolate Salmonella typhi from various water sources and identification of drug resistant genes cat P and tem using Multiplex PCR technique. Enteric fever is prevalent world over and continues to be a major public health problem in developing countries. Infection with Salmonella typhi, the causative organism of this disease, requires effective antimicrobial chemotherapy in order to reduce mortality. Antibiotic-resistant strains of Salmonella are now encountered frequently and the rates of multidrug-resistance have increased considerably in recent years. In the present study, 30 samples were collected from various water sources, of which Salmonella typhi was isolated from 12 samples. All the isolates were subjected to antimicrobial susceptibility testing by standard disc diffusion method against 10 antibiotics. Isolates resistant to chloramphenicol and ampicillin were selected and the drug
resistant genes were identified as cat P and tem respectively by Multiplex PCR technique. There was a very good correlation between the genotypic analysis by PCR and the phenotype determined by standard methods of susceptibility testing.
Keywords: Multiple drug resistance, Salmonella typhi, Disc diffusion technique, DNA extraction, Agarose gel electrophoresis, Multiplex PCR
Full Text:
INTRODUCTION
Water meant for human consumption should be free from pollution and should be safe and acceptable. Indeed the microbial quality of potable water should not exceed limits specified in the water quality guidelines (APHA – 1998).1 The presence of bacteria and pathogenic (diseasecausing) organisms is a concern when considering the safety of drinking water.2 Human and animal wastes are the primary source of bacteria in water. These sources of bacterial contamination include runoff from feedlots, pastures, dog runs, and other land areas where animal wastes are deposited.3,4 Additional sources include seepage or discharge from septic tanks, sewage treatment facilities and natural soil or plant bacteria. Bacteria from these sources can enter wells that are either open at the land surface, or do not have watertight casings or caps. Pathogenic organisms can cause intestinal infections, dysentery, hepatitis, typhoid fever, cholera, and other illnesses.5
Salmonella typhi is the primary aetiological agent of typhoid fever which is responsible for significant morbidity and mortality, particularly in the developing countries. S. typhi is an obligate human pathogen. It causes infection by the faeco-oral route. Typhoid fever is typically acquired by ingesting food or water that has been contaminated by faeces of typhoid-infected individuals. Many outbreaks, caused either by consumption of contaminated water or food have been report.6 Effective antimicrobial therapy is required for the treatment of typhoid fever to reduce morbidity and mortality. Historically, the drugs of choice were chloramphenicol, ampicillin, and co-trimoxazole. Antibioticresistant strains of Salmonella are now encountered frequently and the rates of multidrug-resistance have increased considerably in recent years. Chloramphenicol resistance is known in Salmonella Typhi since 1972, when plasmids of incompatibility group Inc H, coding for chloramphenicol resistance were found in S. typhi. Multi drug resistance, (defined as resistance to all the first line antibiotics used to treat typhoid fever, i.e chloramphenicol, ampicillin, cotrimoxazole and tetracycline) has been endemic in India since 1984. MDR S. typhi have emerged as the newer challenges to treatment of typhoid fever.7-10 So in this study ,we have made an attempt to isolate Salmonella typhi from the water samples collected from in and around Chennai and to identify the resistant gene ,responsible for resistance to the conventional drug therapy of typhoid,which is a life threatening endemic infection.
MATERIALS AND METHODS
A total of 30 water samples were collected from various water bodies like sewage, stagnant, marine, well and ponds from different areas in and around Chennai. Six different samples were collected from above mentioned water sources and it was brought to the laboratory for further processing.
ISOLATION OF SALMONELLA TYPHI
For the isolation of Salmonella typhi, 1ml water samples were inoculated in 9ml selenite-F-broth and incubated for 18hrs at 37oC for enrichment. The enriched samples were plated onto Salmonella-Shigella Agar (Oxoid) and incubated for 24hrs at 37oC. Small colorless colonies suggestive of S.typhi were subjected to preliminary tests like Gram staining and motility and biochemical reactions like Indole, Methyl red, Voges prausker, Citrate utilization, Urease test, Triple sugar iron test and further confirmed by slide agglutination test with specific antisera.10,11 The confirmed colonies of Salmonella typhi were stored at 40C in nutrient agar slant.
ANTIBIOTIC SUSCEPTIBILITY TESTING
The antibiotic susceptibility pattern of the isolated stains were found by disc diffusion technique [Kirby Bauer method]12, against the following antibiotics like Ampicillin(A30mcg/disc), Chloramphenicol(C30 mcg), Tetracycline(TE 30 mcg), Co-trimoxazole( CO 23.75 mcg), Ciproflaxacin(CIP 5mcg), Gentamycin(G 10 mcg), Ceftriazone (Ci 30 mcg) ,Nalidixic acid (Na 30 mcg), Nitrofurantoin (NIT 300 mcg), Amoxycillin (AMC 30mcg). The antibiotic discs were purchased from Hi- media laboratories Pvt limited, Mumbai. Broth culture of the bacterial strains compared to Mac Farland?s standard 11,13 0.5 were prepared. Lawn culture of the test organisms were made on the Muller Hinton agar [MHA-Hi media M1084] plates using sterile cotton swab and the plates were dried for 15 minutes. Filter paper discs impregnated with different antibiotics were placed on the plates. The plates were incubated at 37°C overnight and the zone of inhibition of growth was measured in millimeter diameter.
EXTRACTION OF DNA
Overnight broth cultures of Salmonella typhi was taken for DNA extraction following manufacturer?s instruction using LTRC kit MSS 12. The broth culture was centrifuged at 10,000 rpm for 10 minutes. The supernatant was discarded and the pellet obtained after centrifugation was used for DNA isolation. The pellet was suspended in 300µl of solution A ( 200 µl 1 x TE buffer and of SDS) at room temperature and vortexed completely. It was then incubated at 60° C for 20 minutes and then solution B (300 µl of Phenol: Chloroform: Isoamyl alcohol) was added and completely vortexed. It was centrifuged at 10,000 rpm for 10 minutes. About 500µl of the aqueous supernatant solution was taken in a fresh vial and an equal volume of isopropanol was added and mixed throughly by inverting the vials. It was again kept for centrifugation at 10,000 rpm for 10 minutes. About 200µl of ethyl alcohol was added to the vial and mixed by inverting the tube till the white strands of DNA precipitation were seen. It was then centrifuged at 10000 rpm for 10 minutes and the supernatant was discarded. Then the alcohol was decanted without dislodging the pellet. It was completely air dried to remove ethyl alcohol smell from the vial. To the final pellet, about 20µl of TE buffer was added and mixed by tapping the tube till the solution settle at the bottom. The isolated DNA was separated and visualized with the help of agarose gel electrophoreses and viewed in the UV transilluminator. 15,16
AGAROSE GEL ELECTROPHORESIS
Agarose gel electrophoresis is the easiest and commonest way of separating and analyzing DNA. The DNA is visualized in the gel by addition of ethidium bromide. This binds strongly to DNA by intercalating between the bases and is fluorescent.17 1% agarose gel, was prepared by weighing and mixing1g agarose in 100ml of 1 x TBE buffer in a conical flask. The solution was heated until the agarose was completely dissolved. The gel casting tray was prepared by sealing ends of gel chamber with tape and appropriate number of combs were placed in gel tray. 5 ul of ethidium bromide was added to cooled gel and poured into gel tray and allowed to cool for 15-30 min at room temperature. The combs were removed and the gel is placed in electrophoresis chamber and covered with TAE buffer. 20 µl of digested DNA was loaded along with 3 µl of gel loading buffer onto gel and electrophoresed at 100V for 1 h. The DNA bands were visualized using UV transilluminator.18,19
MULTIPLEX PCR
The multiplex PCR assay offers a rapid, simple and accurate identification of antibiotic resistance profiles and could be used in clinical diagnosis as well as for the surveillance of the spread of antibiotic resistance determinants in epidemiological studies. 20,21 . The protocol developed by Asma Haque et al., 2005 was followed for the amplification of DNA. 22 The procedure was carried with the following PCR temperature cycling parameters: Initial denaturation at 95°C for 45 sec followed by 35 cycles of denaturation at 95°C for 45 s; Primer annealing at 51°C for 45 s, primer extension at 72°C for 1 min and 30 s and the final extension at 72°C for 10 min. After the reaction, 5 µl of loading dye was added to amplify PCR products and mixed well. Then, 20 µl of total sample was loaded on 1% Agarose Gel Electrophoresis.
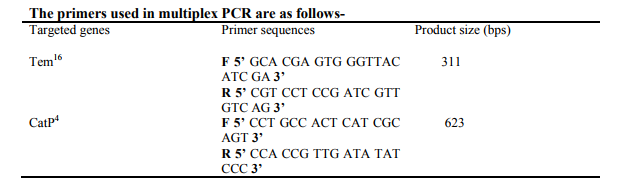
RESULTS AND DISCUSSION
A total of 30 samples were collected from various water bodies. Out of this, Salmonella typhi was isolated from 12 samples. The isolated organisms were confirmed by subjecting to preliminary and various biochemical tests, the results of which are given in table I. Confirmed organisms were subjected to antibiotic susceptibility testing using disc diffusion technique and the zone of inhibition was measured in mm diameter. The results are interpreted as sensitive(S) and resistant(R) and given in table II. Out of 12 isolates, six were resistant to chlorampenicaol. Among this, six strains resistant to chloramphenicol, five were resistant to ampicillin also. The DNA from the samples was extracted and bands, were observed by performing Agarose gel electrophoresis. The isolated DNA was used as template in the PCR study. The protocol developed by Asma Haque et al., 2005 was followed for the amplification of DNA. Multiplex PCR was employed to generate genomic amplification products of Salmonella species. The results showed that there was a very good correlation between the genotypic analysis by PCR and the phenotype determined by standard methods of susceptibility testing and identification of salmonella species:
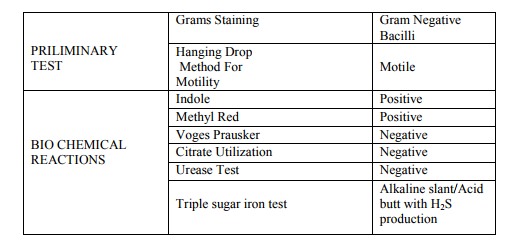
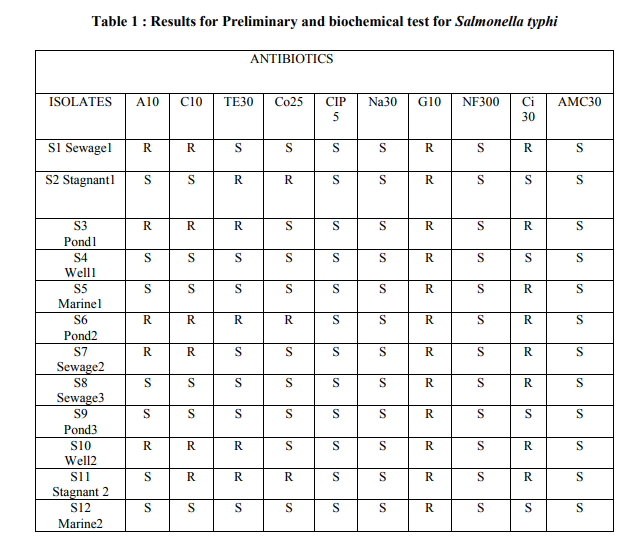
Table II : Results of Antibiotic susceptibility testing of Salmonella typhi A 30- Ampicillin, C 30- Chloramphenicol, TE 30- Tetracycline, CO 25- Co-trimoxazole, CIP 5- Ciproflaxacin, , Na 30- Nalidixic acid, G 10- Gentamycin, NF300- Nitrofurantoin , Ci30- Ceftriazone , AMC 30- Amoxycillin.
Salmonellae are primarily intestinal parasites of humans and many other animals. They are found frequently in sewage, river and other waters, and soil. The presence of Salmonella in other habitats like water, food, natural environment is due to faecal contamination. Under suitable environmental conditions, they may survive for weeks in waters and for years in soils. The prevalence of salmonellosis depends on the water supply, waste disposal, food production and preparation practices, and climate.
Treatment with an appropriate antibiotic is essential for Salmonellosis. Multi drug resistant S. typhi are now endemic in many developing countries and are responsible for significant morbidity and mortality. As a result of the proliferation of such strains, the use of chloramphenicol has been compromised, and that of ampicillin and trimethoprim similarly impaired. Our present study was to isolate Salmonella typhi from various water sources. Isolates resistant to chloramphenicol and ampicillin were selected and the drug resistant genes were identified as cat P and tem respectively by Multiplex PCR technique.
CONCLUSION
Though public health, sanitation and vaccines do have a role to play in control of typhoid fever, it is the antimicrobial therapy which plays a key role in management of typhoid fever. It is likely that S. typhi will continue to acquire genes responsible for antibiotic resistance. Restrictions on the irrational use of antibiotics, and public awareness activities should be undertaken to alert the public to the risks of the unnecessary use of antibiotics .The timely detection of these genes and prevention of their spread is the need of the hour to control antibiotic resistance among S. typhi. More research could also be directed towards factors responsible for their spread and means to prevent dissemination of such plasmids so as to limit drug resistance.
ACKNOWLEDGEMENT
The authors are grateful to Dr.I Seetha Lakshmi,Director of Life Tech Research Centre, Chennai-26 and the Principal ,Valliammal Ammal College for Women,Chennai-102 for their kind support to carry out this project work.
References:
1. APHA (1998). Standard Methods for the Examination of Water and Wastewater, 20th edition. American Public Health Association, Washington.
2. Dufour, A. and Ballentine, R. (1986). Ambient Water Quality Criteria for Bacteria 1986. USEPA, Washington DC
3. Jim Wright, Stephen Gundry and Ronan Conroy, Household drinking water in developing countries: a systematic review of microbiological contamination between source and point-of-use. Tropical Medicine and International Health volume 9 no 1 pp 106–117 January 2004
4. Health Risks From Microbial Growth and Biofilms in Drinking Water Distribution Systems U.S. Environmental Protection Agency Office of Ground Water and Drinking Water Standards and Risk Management Division 1200 Pennsylvania Ave., NW Washington DC 20004 [2002]
5. J.A. Cotruvo, A. Dufour, G. Rees, J. Bartram, R. Carr, D.O. Cliver, G.F. Craun, R. Fayer, and V.P.J. Gannon. World Health Organization (WHO). Waterborne Zoonoses: Identification, Causes and Control.Published by IWA Publishing, London, UK.
6. S Kumar, K Balakrishna, G.P. Singh and H.V. Batra ,Rapid detection of Salmonella typhi in foods by combination of immunomagnetic separation and polymerase chain reaction World Journal of Microbiology and Biotechnology Volume 21, Number 5, 625-628.
7. Threfall EJ, Antimicrobial drug resistance in Salmonella: problems and perspectives in food- and water-borne infections. FEMS Microbiol Rev. 2002 Jun;26(2):141-8
8. Shrikala Baliga. Drug Resistance in Salmonella Typhi: Tip of the Iceberg Online J Health Allied Scs.2004;4:1
9. Cooke FJ, Wain J. The Emergence of Antibiotic Resistance in Typhoid Fever. Travel Medicine and Infectious Disease 2004; 2:67-74
10. Collins,CH and Lyne, P.M 1976.Microbiological methods, London, Butterworths and co.288p
11. J.G.Colle., A.G.Faster., B.P.Marmion., A.Simmons. Practical Microbiology [Mackie and Mc Cartney] 14th edition page 851 – 85
12. Betty A.Forbes., Daniel F.Sahm., Alice S.Weissfeld. Bailey and Scott?s Diagnostic Microbiology 11th edition Mosby page 229 – 257
13. Connie R.Mahon., George Manuselis., Saunder?s Diagnostic Microbiology 2nd Edition
14. L.P. Samaranayake.,Brain M.Jones., Essential Microbiology for Dentistry. Second edition page 217 – 223.
15. Rui Huang, Shuyan Wu, Xueguang Zhang and Yanyun Zhang.,Molecular Analysis and Identification of Virulence Gene on pRST98 from Multi-Drug Resistant Salmonella typhi Cellular and Molecular Immunology, Volume 2 Number 2 April 2005
16. Alessandra Carattoli, PlasmidMediated Antimicrobial Resistance in Salmonella enterica. Curr. Issues Mol. Biol. (2003) 5: 113-122.
17. Sambrook J, Russel DW (2001). Molecular Cloning: A Laboratory Manual 3rd Ed. Cold Spring Harbor Laboratory Press. Cold Spring Harbor, NY. 18. Brody, J.R., Kern, S.E. (2004): History and principles of conductive media for standard DNA electrophoresis. Anal Biochem. 333(1):1-13.
19. David Freifelder, Molecular Biology 2 nd edition 2006:pg 72-76
20. Ali Karami, Zeynab Ahmadi, Zahra Safiri, Fateme Pourali, Detection of Salmonella strain by rapid-cycle multiplex PCR , Junidishapur journal of microbiology (2011); 4(2): 91-98.
21. Cheng-Hsun Chiu And Jonathan T. Rapid Identification Of Salmonella Serovars In Feces By Specific detection Of Virulence Genes, Inva And Spvc, By An Enrichment broth Culture-Multiplex PCR Combination Assay, Journal Of Clinical Microbiology, Oct. 1996, P. 2619– 2622 Vol. 34, No. 10
22. Asma Haque, Abdul Haque, Yasra Sarwar, Aamir Ali, Saira Bashir, Mushkoor Moshin.2010. Identification of drug resistance genes in cinical isolates of Salmonella typhi for development of diagnostic Multiplex PCR. Medical Sciences,vol.21,N(4),P.402-407.
23. Deepa Anbazhagan etal., Multiplex polymerase chain reaction (PCR) assays for the detection of Enterobacteriaceae in clinical samples, African Journal of Microbiology Research Vol. 4(11), pp. 1186-1191, 4 June, 2010
|






 This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License