IJCRR - 7(10), May, 2015
Pages: 55-60
Print Article
Download XML Download PDF
A COMPARATIVE STUDY OF OXACILLIN SCREEN AGAR, OXACILLIN DISC DIFFUSION AND CEFOXITIN DISC DIFFUSION, OXACILLIN E-TEST METHOD FOR ROUTINE SCREENING OF METHICILLIN RESISTANT STAPHYLOCOCCUS AUREUS
Author: Anamika Vyas, Megha Sharma, Sanjeev Kumar, Mrityunjay Kumar, Sudhir Kumar Mehra
Category: General Sciences
Abstract:Background: Methicillin resistant staphylococcus aureus (MRSA) has been recognized as one of the major pathogen in both hospital and community settings. MRSA strains are frequently resistant to different class of antibiotics. Multi drug antimicrobial resistance among MRSA is a matter of concern for clinicians. Therefore, an accurate detection of MRSA in microbiology laboratory is essential for patient management and epidemiological purpose including hospital infection control. Aim: The present study was undertaken to compare various phenotypic methods (oxacillin disc diffusion, cefoxitin disc diffusion, oxacillin screen agar) for detection of MRSA using E test MIC oxacillin as gold standard method. We also aimed to study the resistance pattern of the MRSA isolates. Materials and Methods: A total of 50 staphylococcus aureus strain which were isolated from different clinical specimens were included in this study. All isolates were tested for methicillin resistance by oxacillin disc diffusion, cefoxitin disc diffusion and oxacillin screen agar test considering E test MIC for oxacillin as gold standard. All the isolates were tested for antibiotic susceptibility testing by kirby bauer disc diffusion method against a predefined panel of antimicrobials and intepretation was done according to CLSI guidelines. Result: Among the 50 staphylococcus auresus isolates 23 (46%) isolate were identified as MRSA by E test MIC method. Cefoxitin disc diffusion test showed 100% sensitivity and 92% specificity while oxacillin disc diffusion test and oxacillin screen agar test showed 100% sensitivity and 74% specificity. The resistance percentage of MRSA isolate to erythromycin, ciprofloxacin, levofloxacin, cotrimoxazole and gentamycin was 70%, 96%, 57%, 52% and 43% respectively. All isolates were sensitive to vancomycin, linezolid and tigecycline. Conclusion: Our study revealed that cefoxitin disc diffusion test had high sensitivity and high specificity as compared to other phenotypic methods used routinely to detect MRSA. This method is technically less demanding even can be used along with antibiotic sensitivity testing, cost effective and can be the best option to detect MRSA in clinical settings with constraint facilities. Vancomycin is still the drug of choice for treatment of MRSA, However regular monitoring of vancomycin sensitivity should be done as reduced susceptibility to vancomycin has been reported from all over the globe and is a matter of concern for clinicians.
Keywords: MRSA, Cefoxitin disc diffusion, Oxacillin disc diffusion
Full Text:
INTRODUCTION
Methicillin Resistant Staphylococcus aureus (MRSA) was first discovered in U.K. in 1961 soon after the introduction of methicillin into the clinical practice. Since then MRSA have spread throughout hospitals and other chronic health care facilities worldwide, to the extent that it is now the most commonly isolated antimicrobial resistant pathogen in many countries.1,2 The incidence of MRSA in India ranges from 30-70%.3,4 Traditionally, most strains of MRSA were isolated from hospitalized patients, However MRSA have now appeared in the community world wide in patients with or without risk factor for MRSA infections suggesting a changing epidemiology.5 The importance of MRSA as a nosocomial as well as community acquired pathogen is well documented.6,7 Methicillin resistance in S. aureus is based on production of an additional penicillin binding protein, PBP2 or PBP2a, which is encoded by mecA gene.8 mecA gene is an additional gene found in methicillin resistant S.aureus with no allelic equivalent in methicillin susceptible S. aureus. The major problem in routine screening of MRSA is the heterogeneous population of MRSA. This heterogeneous expression of methicillin resistance in sub population of MRSA can occasionally result in minimum inhibitory concentration that appears to be borderline and consequently the isolate may be interpreted as susceptible.9 Errors in the detection of methicillin resistance can have serious adverse clinical consequences as false susceptibility may result in treatment failure and increased nosocomial and community spread of this deadly microbe if infection control practices are not followed meticulously, on other hand false resistance may not only increase health care cost following unnecessary isolation precautions and over use of glycopeptides but also leads for emergence of clinical isolates with reduced susceptibility to vancomycin. Another major concern about MRSA is that these isolates are frequently resistant to many different classes of antibiotics.10 thus limiting the treatment options to fewer and expensive antibiotics like vancomycin, linezolid and tigecycline. Hence, an accurate identification of MRSA by microbiology lab is essential for institution of effective antimicrobial therapy, infection control measures, epidemiological purpose, and for provision of cost effective health care facilities. Detection of the mecA gene by PCR is the gold standard for identifying MRSA. . However this is a costly and time consuming method and use of this assay is restricted to reference centre and is not routinely carried out in all laboratories.12 Several studies have reported different phenotypic methods developed for detection of MRSA which are widely used in clinical microbiology laboratories.11,12 but the optimum method for the detection remains controversial. Most of the laboratories uses oxacillin disc diffusion method as a routine test for MRSA detection. Cefoxitin,a cephamycin, is potent inducer of mecA regulatory system than oxacillin therefore it is considered better than oxacillin for detection of heterogeneous MRSA. In the present study, we evaluated methicillin resistance in S.aureus isolates by three different phenotypic methods namely oxacillin disc diffusion method, cefoxitin disc diffusion method, oxacillin screen agar method considering E test MIC (oxacillin) as a gold standard . The sensitivity and specificity of each test was determined with the aim to find out a cost effective and easily applicable method or combination there of, for detection of MRSA in a routine diagnostic laboratory. We also aimed to study the resistance pattern of MRSA isolates.
MATERIALS AND METHODS
Study design: This prospective study included 50 staphylococcus aureus strains which were isolated from various clinical specimens submitted to microbiology department of Geetanjali Medical College and Hospital, Udaipur. The specimens included were pus, swabs from surgical wounds, pleural fluid, ascitic fluid, CSF ,Urine, sputum, endotracheal aspirate and blood etc. No duplicate clinical isolates from the same patient and no environmental isolates were included in the study.
Isolation and identification of staphylococci from clinical specimen: All the clinical specimens were first inoculated on Blood agar and McConkey agar plates (Hi media Mumbai,India). Plates were incubated at 37 degree centigrade for 18-24 hrs. S.aureus was identified and differentiated from related organisms on the basis of colony morphology, gram stain, catalase test, slide and tube coagulase test and mannitol fermentation.13
Antibiotic susceptibility testing by Kirby Bauer disc diffusion method: Antibiotic susceptibility testing was performed for all the S.aureus isolates against a predetermined panel of antibiotics by Kirby Bauer disc diffusion method on Muller-Hinton agar plates and the results were interpreted according to the guidelines of the CLSI.14 The antibiotics which were tested included Penicillin(10u), Gentamycin (30µg), Erythromycin(15µg), Clindamycin (2µg), Cotrimoxazole (1.25/23.70µg), Ciprofloxacin (5µg), Levofloxacin (5µg), Vancomycin (30µg), Linezolid(30µg), Tigecycline(15µg), Tetracycline(30µg), Rifampicin (5µg), Chloremphenicol(30µg). S.aureus ATCC25923 was used as a control strain.
Detection of Methicillin resistance by pheno-typic methods: All the S. aureus isolates were tested for methicillin resistance by oxacillin disc diffusion test, cefoxitin disc diffusion test and oxacillin screen agar test. MIC for oxacillin was determined with the E test – strips (Hi-Media Mumbai), which was used as a gold standard method in the present study. The oxacillin disc diffusion test: The oxacillin disc (1µg) diffusion test was carried out on Muller-Hinton agar plates which were supplemented with 2% NaCl to detect MRSA according to the CLSI guideline.14 For each strain a bacterial suspension adjusted to 0.5 McFarland was used. The plates were incubated at 35°C and the results were recorded after 24 hrs. of incubation. The isolates were considered as resistant when the diameter of inhibition was ≤ 10mm, as intermediate resistant when diameter was 11-12mm and as sensitive when the diameter was ≥ 13mm.14
The cefoxitin disc diffusion test: All the isolates were subjected to cefoxitin disc diffusion test using a 30µgm disc. A 0.5 McFarland standard suspension of the isolate was made and lawn culture was done on MHA plates. Plates were incubated at 37°C for 18 hrs. and zone diameter was measured. An inhibition zone diameter of ≤ 21mm was reported as methicillin resistant and a diameter of ≥ 22mm was considered as methicillin sensitive.14 The oxacillin screen agar test: The test was performed by inoculating a direct colony suspension (0.5 McFarland standard) with a swab spotting an area of 10- 15mm in diameter on MHA Plate containg 4% NaCl and 6 µgm/ml oxacillin. Plates were incubated at 35°C for 24 hrs. The plates were observed carefully in transmitted light for any growth. Any growth after 24 hrs was interpreted as oxacillin resistant.15
Determination of MIC by E-Test: MIC for oxacillin was determined with the E-Strip (Hi-media,Mumbai, India) using 0.5 McFarland inoculum according to manufacturers instruction. MHA plates supplemented with 2% NaCl were used. By using cotton swab a lawn culture of standardized bacterial suspension was done on MHA plate. Oxacillin E-strip was then placed on plate and plate was kept in incubation at 35° for 24 hrs. After incubation formation of elliptical zone of inhibition growth occurs. MIC was read where the ellipse intersect the MIC scale on the strip. According to the CLSI standards, S. aureus isolate with oxacillin MIC of ≤ 2µg/ml and ≥ 4µg/ml are defined as methicillin susceptible Staphylococcus aureus (MSSA) and methicillin resistant Staphylococcus aureus (MRSA). MHA plates without antimicrobial were used as control of bacteria growth. S. aureus ATCC 25923 was used as control strain. E-Test MIC was our gold standard method in present study and sensitivity and specificity of other methods were compared with it.
RESULTS
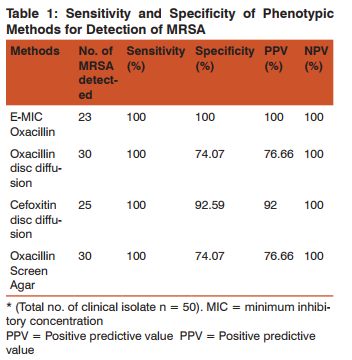
Among the 50 S.aureus isolates 23 (46%) were identified as MRSA by E-Test MIC method. Of 23 MRSA isolates, 16 (69%) strains were isolated from pus, 2 (9%) from urine, blood, and fluids each and 1 (4%) from sputum. Methicillin resistance was detected by oxacillin disc diffusion, cefoxitin disc diffusion and oxacillin screen agar test in 30, 25, 30 isolates respectively. The sensitivity, specificity and the positive and negative predictive values of various phenotypic methods in comparision to E-Test MIC (gold standard), for the detection of MRSA, are Summarized in (Table-1)
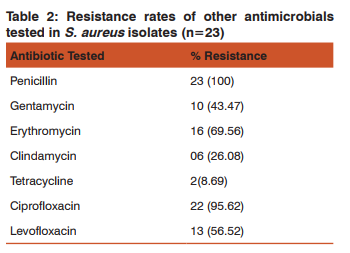
The result of antibiotic resistant rates of MRSA isolates to various antibiotics are shown in table-2. In our study all the strains were sensitive to vancomycin, linezolid,tigecycline, rifampicin and chloremphenicol.

DISCUSSION
Despite the introduction of effective antimicrobial agents and improvements in infection control measures specially hand hygiene, staphylococcus aureus has persisted as important hospital and community pathogen causing superficial skin and soft tissue infections to serious systemic infection leading to illness and death of a person. This problem is further compounded by development of methicillin resistance. Resistance to this antibiotic implies resistance to all β-lactam antibiotics including cephalosporins and monobactams, the most important group of antibiotics to treat staphylococcal infection. Infection with MRSA strains has not only caused therapeutic problems in hospital but also put a tremendous pressure on resources controlling their spread. Thus it is important that clinical microbiology laboratories identify the organism accurately. This will help in determining the appropriate antimicrobial therapy ,shortens the hospital stay, lower hospital cost (by preventing unnecessary use of glycopeptides and isolation precautions), prevent cross transmission in wards and thus in turn will decreases morbidity and mortality. Polymerase chain reaction (PCR) for amplification of the mecA gene is presently considered as the gold standard for detecting methicillin resistance in S.aureus. Inspite of growing consensus in the literature for this method, it is not yet available in all clinical laboratories due to financial and technical constraints, therefore phenotypic methods, although dependent on many environmental and conditional factors still remains a method of choice in resource constraint laboratories. Our study revealed that, overall rate of methicillin resistance with S.aureus was 46%. Similar isolation rates were found in studies from different parts of India, ranging from 45.36 to 59.3%.16,17,18 In contrast 26.4% and 19.5% prevalence rates has also been reported in some studies.19,20 which is comparatively less then that reported in present study. This discrepancy could be due to difference in the study design i.e. Population under study and geographical distribution ,variation in antibiotic usage and infection control practices in different hospitals as well as due to differential clonal expansion and drug pressure in community. MRSA isolates were predominantly isolated from the pus (69%), similar findings were reported by Anupurba et al.17 and Sasirekha et al.21 In present study E-Test MIC determination for oxacillin was used as a gold standard for MRSA detection. The advantage of E-Test method is that it is easy to perform as a disk diffusion test and approaches the accuracy of PCR for mecA gene. There are many studies comparing E-Test MIC with broth dilution and PCR methods which has yielded satisfactory results.22 In present study cefoxitin disc diffusion was found to be highly sensitive 100% and specific 92.59% while sensitivity of oxacillin disc diffusion was 100% and specificity was 74.07%. Similar results were quoted by several other studies.12,16,23,24 Cefoxitin is a better inducer of the expression of the mecA gene, so the heterogeneous population that variably express the mecA gene is better detected by disc diffusion with cefoxitin then with oxacillin, which is a weak inducer of PBP2a production. Several workers have reported that the result of cefoxitin disc diffusion test co-relates better with the presence of mecA gene than the result of oxacillin disc diffusion test.12,25 There are a number of studies stating that cefoxitin disc diffusion method is a reliable method for detection of MRSA and the result were found to be in concordance with PCR mecA gene detection method.24,26,27. Our study also strengthens the fact that cefoxitin is superior to oxacillin as indicator of MRSA for the detection of methicillin resistance. In our study, oxacillin disc diffusion method was only 74.07% specific. The high false positivity of oxacillin disc diffusion method in present study could be due to hyper production of β-lactamase which may lead to phenotypic expression of oxacillin resistance resulting in a clinical isolate which is oxacillin resistant but do not possess the usual genetic mechanism for such resistance. Probably such strains under antibiotic pressure may eventually turn into fully resistant strain. The oxacillin screen agar medium showed 100%.sensitivity and 74.07% specificity. Similar finding of high sensitivity and low specificity using oxacillin screen agar medium was reported by other workers also.12 Difficulty in MRSA detection by oxacillin screen agar base occur if the organism have their MIC near break points i.e. (borderline resistance strain) . The test also performed less well in studies where hetero resistant strains were included in study group, as it is subjected to many environmental conditions such as temperature, pH, salt concentration, incubation time.26 Swenson et al.15 also noted that sensitivity is decreased when hetero resistant strains were tested and specificity decreased with strains having borderline MIC.
Some of the studies have shown different sensitivity and specificity for these three phenotypic tests for detection of MRSA. Baddour et al.31 reported that the sensitivity and specificity of the cefoxitin and oxacillin disk diffusion test were 84.6%, 84.6%, 87.5% and 79.2% respectively. They found that the oxacillin agar screening was 92.3% sensitive and 45.8% specific. In another study by Jain et al.32 the sensitivity and specificity of the cefoxitin and oxacillin disk diffusion test were 94.44%, 100%, 95.83% and 58.33% respectively. Matos et al.33showed the cefoxitin and oxacillin disk diffusion test and oxacillin agar screening was 100% specific but only the cefoxitin and oxacillin disk diffusion test had 100% sensitivity. They reported that the oxacillin agar screening had the lowest sensitivity (82.2%). In general, in the most conducted studies, cefoxitin disk diffusion test has shown the highest specificity compared to oxacillin disk diffusion and agar screening. In a laboratory where it is not possible to carry out molecular method as a routine, cefoxitin disk diffusion test is a good surrogate marker for detecting methicillin resistance. It is far superior to most of the currently recommended phenotypic method like oxacillin disc diffusion and oxacillin screen agar method. No special medium or incubation temperature is required for cefoxitin as is required for oxacillin and results are easy to read in both transmitted and reflected light. It is now an acceptable method for detection of MRSA by many reference groups including CLSI. Considerable variations were found in the reported resistance profile among MRSA isolates from different countries and from different hospitals with in a country. Keeping in view this fact we determined the resistance pattern of MRSA isolates against a pre-determined panel of antimicrobials. Among MRSA isolates high degree of resistance was encountered for ciprofloxacin (96%), levofloxacin (57%), erythromycin (70%), cotrimoxzole (53%). This is similar to the finding of studies carried out by Sasirekha et al21 and Udo et al30 which also found high level of resistance to erythromycin and ciprofloxacin. The present study revealed high percentage of sensitivity to gentamycin (57%). Similar finding was also reported in a study carried out by Sasirekha et.al.21 This is in contrast to the studies done by Quereshi et al28 and Kandle et al.29 They reported 97.8% and 91% resistance to gentamycin. The reason which could justify these finding is that gentamycin is not used frequently to treat staphylococcus infection in our set up thus decreasing selection pressure for drug resistance, at the same time macrolides and quinolones are broad spectrum antibiotics, frequently used in the treatment of common staphylococcal infection. This change in antibiotic usage pattern would have led to the development of gentamycin sensitive and macrolide, quinolones resistant isolates. In our study no strain was found resistant to vancomycin, linezolid which was similar to other studies.4,11,14,16,17 Sensitivity to tigecycline was also 100%. This may be due to the fact that due to high cost these drugs were not used frequently in our setup thus decreasing the selection pressure for drug resistance. However MRSA strains with reduced susceptibility to vancomycin have been reported recently from various parts of country.
CONCLUSION
It is concluded from the present study that cefoxitin disc diffusion method had a high sensitivity and specificity compared to other phenotypic methods for detection of MRSA. Cefoxitin disc diffusion method can be the preferred option to detect MRSA in clinical settings with resource constraint facilities as it is easy to perform, do not require special technique, media preparation and finally more cost effective than PCR and latex agglutination test for PBP2a detection. Vancomycin, linezolid, tigecycline are effective drugs for treatment of MRSA. We suggest that these drugs should be considered as reserve drugs and should not be used as empirical therapy in treatment of staphylococcus aureus and other gram positive infections. Regular monitoring of vancomycin sensitivity should be carried out to find out early emergence of VISA or VRSA strains in clinical setup. It was noted that MRSA isolates showed resistance to most of the antibiotics. This finding calls, for urgent attention where by strict antibiotic policy should be enforced to curtail irrational use of antibiotics. Constant surveillance of antimicrobial profile of MRSA isolates should be carried out which will help the clinicians for selection of appropriate antimicrobial therapy.
ACKNOWLEDGEMENT
Authors acknowledge the immense help received from the scholars whose articles are cited and included in references of this manuscript. The authors are also grateful to authors/editors/publishers of all those articles, journals and books from where the literature for this article has been reviewed and discussed.
References:
1. Diekema DJ, BootsMiller DJ, Vaughn TE, Woolson RF, Yankey JW. Antimicrobial resistance trends and outbreak frequency in United States hospitals. Clin. Infect. Dis.2004; 38:78-85.
2. Goosens H. European Status of resistance in nosocomial infections. Chemotherapy.2004; 51:177-181.
3. Verma S, Joshi S, Chitnis V, Hemwani N, Chitnis D. Growing problems of Methicillin Resistance Staphylococci-Indian Scenario Indian J. Med Sci 2000;54(12):535-40.
4. Rajaduraipandi K, Mani KR, Panneerselvam K, Mani M, Bhaskar M and Manikandan P. Prevalence and antimicrobial susceptibility pattern of methicillin resistant Staphylococcus aureus: A multicentre study. Indian J. Med. Microbiol 2006;24(1):34-8.
5. Chambers HF. The changing epidemiology of staphylococcus aureus Emerg. Infec. Dis.2001;7:178-182.
6. Brown DF, Edward DI, Hawkey PM, Morrison D, Ridgway GL, Towner KJ, et.al. Guidelines for the laboratory diagnosis and susceptibility testing of methicillin-resistant staphylococcus aureus (MRSA). J. Antimicrob chemother 2005; 56(6):1000-18
7. Rabhar M, Yaghoobi M, Fattahi A.Comparision of different laboratory methods for detection of Methicillin Resistant Staphylococcus aureus.Pak J med Sci. 2006;22(4):442-5.
8. Brown D.F. Detection of methicillin/oxacillin Resistance in staphylococci. J Antimicrob chemother. 2001;48 suppl 1:65-70.
9. Swenson JM, Patel JB, Jorgensen JK. Special phenotypic methods for detecting antibacterial resistance, chapter 74. In:Murray PR, Baron ES, Jorgensen JH, Landry ML Pfaller MA, editors. Manual of Clinical Microbiology. 9th ed. Washington DC:ASM Press 2007 P.1175-76.
10. Tiemersma EW, Bronzwaer SL, Lyytikainen O, Degener JE, Schrijnemakers P, Bruinsma N, et.al. Methicillin-resistant staphylococcus aureus in Europe, 1999-2002. Emerg. Inf. Dis. 2004; 10(9):34-7.
11. Tiwari HK, Sapkota D, Das AK, Sen MR. Assesment of different methods to detect Methicillin Resistnt Staphylococus aureus. Southeast Asian J.Trop Med Public Health 2009;40:801-06
12. Velasco D, Mar Tomas MD, Cartelle M, Beciero A, Perez A, Molina F et al Evaluation of different methods for detecting methicillin (oxcillin) resistance in staphylococcus aureus. J. Antimicrob Chemother 2005; 55(3):379-82.
13. Baird D, Staphylococcus : Cluster forming gram positive cocci. In:Mackie and McCartney Practical Medical Microbiology, Colle J.G, Fraser A.G., Marmion BP., Simmons A, (14th Ed) Churchill Livingstone, 1996, PP:245-261.
14. Clinical Laboratory Standard Institute. Performance standard for Antimicrobial susceptibility testing ; twentieth Informational supplement CLSI documentM100-S20.Wayne, PA:CLSI;2009
15. Swenson J.M., Williams, P.P., Killgore, G., O’Hara C.M. and Tenover FC . Performance of eight methods, including two new rapid methods, for detection of oxacillin resistance in a challenge set of staphylococcus aureus organisms J. Clin Microbial.2001; 39: 3785-3788.
16. Oberoi L, Kaur R. and Aggarwal A. Prevalence and antimicrobial susceptibility pattern of Methicillin Resistant Staphylococcus aureus (MRSA) in a rural tertiary care hospital in North India . IJABPT 2012;3(1):200-05.
17. Anupurba S, Sen MR, Nath G, Sharma BM, Gulati AK, Mohapatra T.M. Prevalence of methicillin resistant Staphylococcus aureus in tertiary referral hospital in Eastern Uttar Pradesh, Indin J. Med Microbial 2003;21(1):49-51.
18. Tiwari H.K, Sen M.R. emergence of vancomycin resistant Staphylococcus aureus (VRSA) from a tertiary care hospital from northern part of India. BMC Infect Dis 2006;6:156.
19. Kumari N, Mohapatra TM, Singh YI. Prevalence of Methicillin Resistant Staphylococcus aureus (MRSA) in a tertiary - care hospital in Eastern Nepal. J. Nepal Med Assoc 2008; 47(170):53-56.
20. Tahnkiwale SS, Roy S, Jalgaonkar SV Methicillin reistance among isolates of staphylococcus aureus: Antibiotic sensitivity pattern and phage typing. Ind J Med Sci 2002;56:330- 334.
21. Sasirekha B, Usha M.S., Amruta A.J., Ankit S, Brinda N, Dviya R. Evaluation and Comparision of different phenotypic tests to detect Methicillin Resistant Staphylococcus aureus and their Biofilm Production. Int J.PharmTech Res.2012 ; 4(2) : 532-541.
22. Ercis S, Sancak B, Hascelik G. A comparision of PCR detection of mec A with oxacillin disk susceptibility testing in different media and sceptor automated system for both staphylococcus aureus and coagulase negative staphylococci isolates. Indian J Med Microbial 2008;26(1):21-24.
23. Tiwari HK, Sapkota D, Das AK and Sen MR. Assesment of different methods to detect Methicillin Resistant Staphylococcus aureus. Southeast Asian J. Trop Med Public Health 2009;40(4):801-06.
24. Mathews A.A., Thomas M., Appalaraju B., Jayalakshmi J., Evaluation and Comparision of tests to detect methicillin Resistant S. aureus. Ind J. Pathol Microbial 2010;53(1):79- 82
25. Cauwelier B, Gordts B, Descheemaecker P, Van Landuyt H, Evaluation of a disk diffusion method with cefoxitin (30µg) for detection of methicillin resistant Staphylococcus aureus Eur. J. Clin Microbial. Infect Dis.2004; 23(5):389-392.
26. Anand K.B., Agarwal P., Kumar S., Kapila K., Comparison of cefoxitin disc dffusion test, oxacillin screen agar, and PCR for mecA gene for detection of MRSA. Ind. J. Med. Microbiol. 200; 27(1):27-29.
27. Rahbar M., Safadel N., Evalution of cefoxitin disk diffusion test for routine detection of methicillin resistant Staphylococcus aureus. Iran J. Patholol.2006;1(4):145-148.
28. Qureshi A.H., Rafi S., Qureshi S.M., Ali A.M., The current susceptibility patterns of MRSA to conventional antistaphylococcal antimicrobials at Rawalpindi. Pak. J. Med. Sci. 2004; 20:361-364.
29. Kandle S.K., Ghatole M.P., Takpare A.Y., Hittinhalli V.B., Yemul V.L., Bacteriophage typing and antibiotic sensitivity pattern of staphylococcus aureus from clinical specimen in and around Solapur (South Maharashtra). J. commun. dis. 2003; 35:17-23.
30. Udo E.E., Sweih A.N., Mokaddas e., Johny M., Dhar R., Gomaa H.H., Obaid I.A., Rotimi V.O., Antibacterial resistance and their genetic location in MRSA isolated in Kuwait hospitals, 1994-2004. BMC Infect. Dis.;2006, 6(1):168.
31.Baddour MM, Abuelkheir MM, Fatani AJ. Comparison of mecA polymerase chain reaction with phenotypic methods for the detection of methicillin-resistant Staphylococcus aureus. Curr. Microbiol.2007; 55: 73-479.
32. Jain A, Agarwal A, Verma RK . Cefoxitin disc diffusion test for detection of meticillin-resistant Staphylococci. J. Med. Microbiol.2008; 57:957-961.
33. Matos PD, Schuenck RP, Cavalcante FS, Caboclo RM, Santos KR . Accuracy of phenotypic methicillin susceptibility methods in the detection of Staphylococcus aureus isolates carrying different SCCmec types. Mem. Inst. Oswaldo. Cruz.2010; 105:931-934.
|






 This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License