IJCRR - 14(3), February, 2022
Pages: 36-42
Date of Publication: 01-Feb-2022
Print Article
Download XML Download PDF
Raspberry Pi (Python AI) for Plant Disease Detection
Author: Shagufta Aftab, Chaman Lal, Suresh Kumar Beejal, Ambreen Fatima
Category: Healthcare
Abstract:The diagnosis of diseases at an early stage is the main goal of this paper. We concentrate on image processing techniques in this research. This entails a range of processes ranging from taking a picture of the leaves to using Raspberry PI to diagnose the condition. The Raspberry PI is used to connect the camera to the display device, from which the data is sent to the cloud. Various procedures, such as acquisition, pre-processing, segmentation, and clustering, are used to examine the acquired images. As a result, the demand for labour in big farm areas is reduced. Also, the cost and effort are reduced, whereas productivity is increased. Various procedures, such as acquisition, pre-processing, segmentation, and clustering, are used to examine the acquired images. As a result, the demand for labour on huge farmlands is reduced. Costs and efforts are also minimized, while production is raised.
Keywords: Raspberry PI, segmentation, Image-processing, Artificial intelligence, Clustering, Disease detection
Full Text:
Introduction
This current paper provides motivation and a brief overview of our study. In terms of monitoring the crops of large farms with minimum staff, presently, technology adoption in farming has shown quantitative outcomes in agricultural productivity. The agriculture sector has been changed by the Internet of Things (IoT), Cloud Computing, Artificial Intelligence, and Computer Vision, which have all helped to boost productivity over time with minimal investment. The failure to diagnose agricultural diseases in their early stages is a key worry that has a negative impact on crop output. In most cases, crop disease identification is done manually. In general, crop disease detection is done by hand, and it is impossible to identify crop illnesses without the help of experts who have acquired knowledge about the signs and causes of the diseases.
The importance of accurate and timely illness detection, as well as early prevention, has never been greater in this changing world. Plant diseases can be detected in a variety of methods. Some diseases have no visible symptoms, or the damage becomes apparent too late to intervene, necessitating a thorough investigation. However, because most diseases exhibit themselves in the visible spectrum, a skilled professional's naked eye examination is the primary method for detecting plant diseases in practice. A plant pathologist must have good observation skills to recognize distinctive symptoms to diagnose plant diseases accurately7. In this regard, an automated system that can identify plant illnesses based on the look and visual symptoms of the plant might be extremely useful to both amateur gardeners and skilled professionals as a disease diagnosis verification system. An automated system that could identify plant illnesses based on the appearance and visual symptoms of the plant might be extremely useful to both amateur gardeners and skilled professionals as a disease diagnosis verification system.
The suggested approach uses machine learning to detect and classify various plant leaf diseases3, 10. There are four primary steps in the plan. The segmentation process begins with the creation of a colour transformation structure for the input RGB image, followed by the masking and removal of the green pixels using a certain threshold value, and finally the segmentation process. For the effective segments, texture statistics are generated, and the retrieved features are then fed to the classifier.
BACKGROUND
There has been a slew of earlier studies on plant categorization using picture data and technologies like Probabilistic Neural Networks (PNN) and Support Vector Machines (SVM). Using image processing techniques15, plant diseases can be detected. A digital camera is used to capture photographs of the plant, which is connected to the Raspberry Pi board4. To obtain the features for further analysis, various image processing techniques are applied to the acquired image13. A series of processes are included in this image processing procedure mentioned below.
-
IMAGE ACQUISITION:
The camera module captures the RGB photos from the plant. Because the camera has a resolution of 21 megapixels, the RGB photographs are quite clear.
-
TRANSFORMING A RGB IMAGE TO HSV FORMAT:
The RGB pictures are transformed to Hue Saturation Value, a colour space that is an excellent tool for colour perception. RGB is used as an ideal representation for colour creation. Like the observer's perseverance, the hue is nothing more than a colour characteristic that expresses pure colour. Saturation, also known as relative purity, is the representation of the quantity of white light added to the hue of the image. The amplitude of light is referred to as its value. The Hue component is included in the analysis, but the Saturation and Value components are excluded because they do not contribute any further information.
-
PIXEL MASKING IN GREEN:
Masking is the process of altering a pixel's background value to zero or any other value in a picture. This step detects the pixels that are mostly green in colour.
4. REMOVING GREEM PIXEL MASKS:
The green pixels are then set to zero based on the provided threshold value computed for the pixels. RGB component mapping assigns a value of zero to the pixel's red, green, and blue components. Because the healthy portions of the leaf are represented by green-colored pixels, they do not aid in disease identification.
5. COMPONENT SEGMENTATION:
The contaminated area of the leaf is excised and split into several equal-sized segments.
6. COLLECTING THE IMPORTANT SECTIONS FROM THE PROCESS IMAGE:
There is no relevant information in any of the portions. For analysis, only segments with a significant amount of data are chosen.
7. COLOR CO-OCCURENCE METHOD:
The texture features are produced from the statistical distribution of observed intensities at specified points in the image.
8. EVALUATE THE TEXTURE STATISTICS:
For the color content of the image, the contrast, local homogeneity, energy, and correlation are computed. The contrast function returns the difference in intensity between a pixel and its neighbors.
RELATED WORK
Plant diseases have a significant impact on the growth of their individual species, hence early detection is essential. Machine Learning (ML) models have been used to detect and classify plant illnesses1,6,9. but with recent advances in a subset of ML, Deep Learning (DL) 2, this area of research appears to have a lot of promise in terms of improved accuracy. To detect and classify the symptoms of plant illnesses, several developed/modified DL architectures 14, as well as many visualization techniques, are used. In addition, these architectures/techniques are evaluated using a variety of performance measures. The DL models used to illustrate numerous plant diseases are thoroughly explained in this article 5, 8. Furthermore, several research gaps have been uncovered, allowing for increased transparency in detecting plant illnesses even before symptoms show.
Laboratory-based tests are used in direct detection approaches. Indirect methods, on the other hand, rely on sophisticated methodologies with a focus on imaging tool integration.
Indirect approaches rely on the on-site integration of sensors and smart devices to give a faster and more accurate method of illness identification. Early detection of apparent plant illnesses is critical, as it allows farmers to take the necessary actions to save the damaged plant. If early detection is possible, the percentage of damaged fruits can be reduced dramatically while still maintaining excellent production standards.
a. DIRECT METHODS
When a pathogen infects a plant, the DNA of the plant is altered, and the pathogen produces and introduces a specific type of protein molecule to the plant. Direct methods use molecular and serological techniques to look for pathogen DNA or pathogen-produced protein molecules in the plant's biological structure. Polymerase chain reaction (PCR) and enzyme-linked immunosorbent assay (ELIA) are two often-used procedures (ELISA). The genetic material (DNA) of the bacteria causing the disease is extracted utilizing PCR-based disease detection. After the DNA has been purified and amplified, it is run through gel electrophoresis. After the DNA has been purified and amplified, gel electrophoresis is carried out. The presence of a specific brand in the gel electrophoresis verifies the existence of the plant disease organism 13.
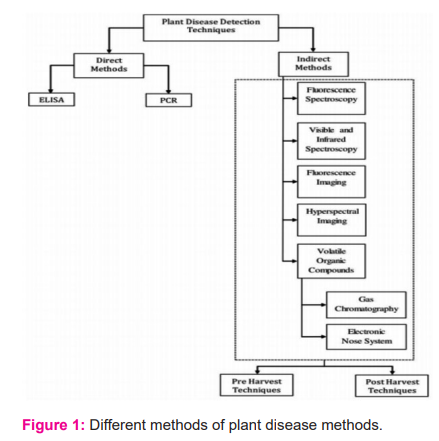
Although these procedures are reliable and accurate in detecting plant diseases [fig: 1], they have several disadvantages. These methods rely largely on expensive laboratory equipment and lengthy experiments, both of which can be time-consuming and labor-intensive. To ensure trustworthy and precise results, sample preparation takes a significant amount of time and work. Because of the usage of consumable reagents that are individually formulated for each pathogen, these procedures are also quite expensive 12. As a preliminary screening tool for processing huge numbers of plant samples, better and faster disease detection technologies are required.
b. INDIRECT METHODS
It was studied whether new automated non-destructive technologies could be used to detect plant disease symptoms early and with high sensitivity to specific diseases. These technologies should be able to detect illnesses and stressors in real-time in the field. The imaging technique is a popular method.
Researchers applied deep learning architectures [fig: 2] to image recognition and classification as they evolved over time 11. These structures have been used in a variety of agricultural applications as well. The performance of an author-modified CNN and Random Forest (RF) classifier was tested through CA at 97.3 percent in the classification of leaves among 32 species.
c. CONVOLUTIONAL NEURAL NETWORK
Using deep learning approaches, convolutional neural network models were constructed to detect and diagnose plant diseases using simple leaf photos of healthy and ill plants. The models were trained using an open collection of 87,848 photos, which included 25 different plants in 58 different classes of [plant, illness] pairs, including healthy plants. Several model architectures were trained, with the top performing one achieving a success rate of 99.53 percent in detecting the corresponding [plant, illness] pair (or healthy plant). The model's high success rate makes it a valuable advising or early warning tool, as well as a technique that might be expanded to support an integrated plant disease diagnosis system that can operate in real-world situations.
system requirement
a. OPEN CV
Open CV is a cross-platform library for developing real-time computer vision apps. It primarily focuses on image processing, video recording, and analysis, including capabilities such as face and object detection. It is critical in real-time operation, which is critical in today's systems. It may be used to process photos and videos to recognize items, faces, and even human handwriting.
b. TENSOR FLOW
The feature extraction network must accurately extract the properties of the disease image to achieve a high disease recognition rate. Tensor flow is used to do this. The convolutional neural network, as a deep learning model, is capable of hierarchical learning and excels at feature extraction. The Tensor flow Object Detection API is used in our project.
Tensor flow’s object detection API provides a framework for building a deep learning network that can tackle object detection challenges. In their framework, which they call Model Zoo, there are already pertained models. This comprises models that have been pre-trained using the COCO, KITTI, and Open Images datasets. If we're solely interested in categories, these models can be employed for inference this collection of information. They're also useful for training on a new dataset and initializing your models.
For a clear image, we employ a 5-megapixel camera that concentrates on plants. It connects to the Raspberry PI 4 board via USB.
METHODOLOGY
a. DATA COLLECTION
The disease's exact location is then retrieved from the image and saved as an XML file.
b. MODEL IMPLEMENTATION
The information is fed into the API, which employs the Mobile-NET SSD (Single Shot MultiBox Detector) feature extractor for real-time detection and the FASTER RCNN for single image detection. The API will iterate over the image until we're happy with the outcome.
c. MODEL TESTING
The model is then tested using testing photos when the training is done.
RESULT AND ANALYSIS
If the user hasn't already logged in, this is the first window that will open [fig: 3]. This has two primary buttons login and signup, which allow users to log in or register to enjoy the system's features.
This is the main graphical user interface [fig: 4], through which the user can choose from a variety of settings.
By clicking on the open picture button, the user can choose a leaf image [fig: 5], as illustrated in figure.

This window will appear if you touch/click the “Run Detection” button. To begin detecting the image you just opened above, press "No" [figure: 6].
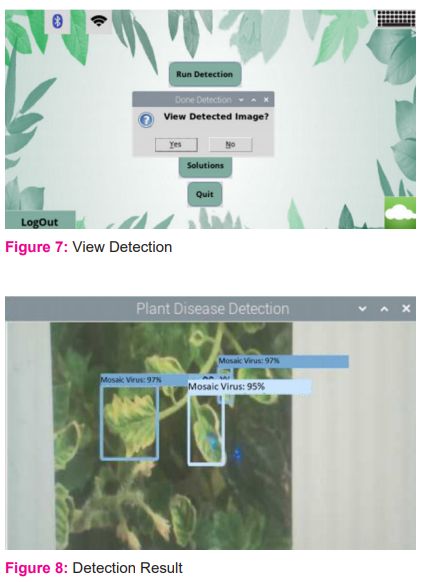
When the detection is finished, you will be asked if you wish to view the image that was detected or not. To see what the system detects, press "Yes" in [figure: 7].
In [figure: 8], shows, the system detects the plant and the type of disease it has.

Small, light to dark green, round to irregularly shaped water-soaked dots are the earliest signs of late blight in the field [fig: 9]. Although the symptoms are similar, the treatment method differs. The steps below should be followed:
Always purchase new seed potatoes that are certified, and disease-free.
Keep developing tubers covered with soil.
Protectant fungicides, like chlorothalonil and fixed copper, can help protect foliage if applied prior to infection.

Small, yellow-green lesions on young leaves that are frequently distorted and twisted, or black, water-soaked, greasy-appearing lesions on older foliage [fig: 10], are the first signs. The steps below should be followed:
Select resistant varieties
Seed and transplants that are disease-free should be purchased. Soak seeds in a 10% chlorine bleach solution for 2 minutes to treat them (1 part bleach; 9 parts water). Before planting, thoroughly rinse and dry the seeds.
Plants should be mulched deeply with a thick organic substance such as newspaper coated in straw or grass clippings.
Avoid overhead watering.
At the conclusion of the season, remove and destroy any sick plant parts as well as all trash.
To inhibit the spread of infection, spray with fixed copper (organic fungicide) every 10-14 days.
If infections are severe, move peppers to a different site and cover the soil with black plastic mulch or black landscape cloth before planting.
These are some of the plant disease detection results.
-
-
DATAFLOW DIAGRAM
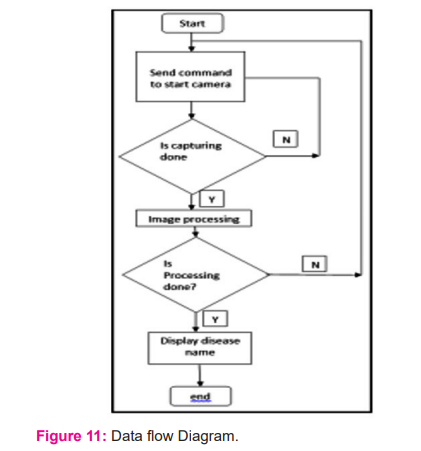
[Figure: 11] shows a data flow diagram that explains the system's operation. When the system starts, it sends a command to capture an image, then image processing begins, and the illness name is shown.
-
-
USE CASE DIAGRAM
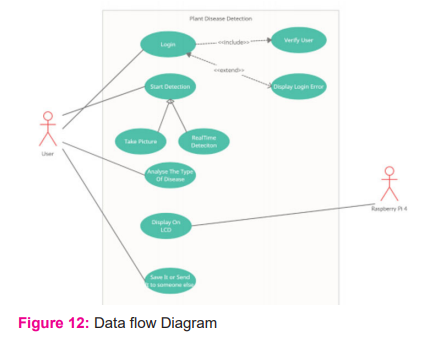
The flow of work is depicted in the diagram [fig: 12]. To access the system's features, the user must be logged in. The system gives the user two choices:
1. To use a photograph to do detection. He or she can take pictures using a pi4 camera.
2. Alternatively, you can employ real-time detection. The sickness will be detected in real-time because of this.
FEATURES OF THE SYSTEM
-
The suggested system uses image processing to detect plant illness.
-
The system is user-friendly; we can quickly obtain data from collected images.
-
The recognized image is saved in the system's database for future usage.
-
The accuracy of the system can be improved by data entry and picture processing of big data sets.
-
With further modifications, this system will benefit society in the future.
-
The system has a 99.99 percent accuracy level.
CONCLUSION
We developed a classification approach for picture-based plant identification and content-based image retrieval challenges in this research. To detect plant diseases, the Tensor flow Object Detection API is employed. Two alternative models, Faster RCNN for improved accuracy and SSD Mobile net for disease detection in real-time, were trained on diverse plant illnesses and their healthy states. By putting our models to the test, we can assess how well they can detect different plant illnesses given a picture. The model's accuracy is also mentioned, which is sufficient for detecting practically every plant disease. Different detections are shown to evaluate the model's accuracy. Tomatoes, potatoes, and bell peppers may all be tested for illnesses with this approach.
ACKNOWLEDGEMENT: Nil
Source of Funding: Nil
Conflict of Interest: Nil
Author’s contribution:
Shagufta Aftab: data collection.
Chaman lal: writing of first and final drafts.
Suresh Kumar B: Manuscript Editing.
Ambreen Fatima: data collection.
References:
1. Shruthi U, Nagaveni V, Raghavendra BK. A review on machine learning classification techniques for plant disease detection. In2019 5th International Conference on Advanced Computing & Communication Systems (ICACCS) 2019 Mar 15 (pp. 281-284).
2. Nagaraju M, Chawla P. Systematic review of deep learning techniques in plant disease detection. Int Journal of Syst Assur Eng Manag, 2020 Jun;11(3):547-60.
3. Deepika P, Kaliraj S. A Survey on Pest and Disease Monitoring of Crops. In2021 3rd International Conference on Signal Processing and Communication (ICPSC) 2021 May 13 (pp. 156-160).
4. Sankaran S, Mishra A, Ehsani R, Davis C. A review of advanced techniques for detecting plant diseases. Computers and electronics in agriculture. 2010 Jun 1;72(1):1-3.
5. Tiwari D, Ashish M, Gangwar N, Sharma A, Patel S, Bhardwaj S. Potato leaf diseases detection using deep learning. In2020 4th International Conference on Intelligent Computing and Control Systems (ICICCS) 2020 May 13 (pp. 461-466).
6. Ahmed K, Shahidi TR, Alam SM, Momen S. Rice leaf disease detection using machine learning techniques. In2019 International Conference on Sustainable Technologies for Industry 4.0 (STI) 2019 Dec 24 (pp. 1-5).
7. Durmu? H, Güne? EO, K?rc? M. Disease detection on the leaves of the tomato plants by using deep learning. In2017 6th International Conference on Agro-Geoinformatics 2017 Aug 7 (pp. 1-5).
8. Durmu? H, Güne? EO, K?rc? M. Disease detection on the leaves of the tomato plants by using deep learning. In2017 6th International Conference on Agro-Geoinformatics 2017 Aug 7 (pp. 1-5).
9. Harish S, Gayathri KS. Smart Home-based Prediction of Symptoms of Alzheimer’s Disease using Machine Learning and Contextual Approach. In2019 International Conference on Computational Intelligence in Data Science (ICCIDS) 2019 Feb 21 (pp. 1-6).
10. Soni H, Arora P, Rajeswari D. Malicious Application Detection in Android using Machine Learning. In2020 International Conference on Communication and Signal Processing (ICCSP) 2020 Jul 28 (pp. 0846-0848).
11. Valdoria JC, Caballeo AR, Fernandez BI, Condino JM. iDahon: An android based terrestrial plant disease detection mobile application through digital image processing using deep learning neural network algorithm. In2019 4th International Conference on Information Technology (InCIT) 2019 Oct 24 (pp. 94-98).
12. Ramesh S, Hebbar R, Niveditha M, Pooja R, Shashank N, Vinod PV. Plant disease detection using machine learning. In2018 International conference on design innovations for 3Cs compute communicate control (ICDI3C) 2018 Apr 25 (pp. 41-45).
13. Kusumo BS, Heryana A, Mahendra O, Pardede HF. Machine learning-based for automatic detection of corn-plant diseases using image processing. In2018 International Conference on Computer, Control, Informatics, and its Applications (IC3INA) 2018 Nov 1 (pp. 93-97).
14. Modem Amarendhar R, M. James S, P.V.G.D Prasad R. Analysis of COVID-19 Complications Using Deep Learning-Based Neuro-Fuzzy Classification Approach. Int J Cur Res Rev. 13(20), October, 2021, 85-89,
15. Nanditha B R, Geetha Kiran A. A Review on Imaging Modalities and Techniques for Oral Malignancy Detection Int J of Cur Res Rev. 13(20), October, 71-78.
|






 This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License