IJCRR - 13(19), October, 2021
Pages: 58-64
Date of Publication: 11-Oct-2021
Print Article
Download XML Download PDF
Tolerance and Remediation Potential of Water Microbes against Nitrate
Author: Thakur Preeti, Gauba Pammi
Category: Healthcare
Abstract:Introduction: Nitrate is the major concern of groundwater. High concentration arises due to the massive application of chemical fertilizers. It causes various issues Blue Baby Syndrome, respiratory problems etc. Conventional methods are available to remove nitrate, but they can sequester the nitrate and not treat it. Bioremediation is a low-cost technique and is efficient in nitrate removal. Objective: Isolation and identification of nitrate-reducing microbes from the polluted site of Yamuna river. Methods: This paper focuses on microbe's isolationusingBioremediation technique and their identification through Molecular and Biochemical tests. Result: 4 microbes were found to have the potential for remediating nitrate. Microbes were inoculated in nitrate broth and incubated (37?C) under both aerobic and anaerobic conditions. Phenotypical and molecular characterization was done. The consensus sequence was submitted in GenBank and accession numbers were obtained.These were Enterobacteraerogenes (MN252552), Lelliotiaamnigena (MN647560), E. coli K12 (MN754025) and Klebsiella oxytoca (MT457847) respectively. After incubation, these strains showed 48.4%, 34.1%, 42.90% and 33.66% of nitrate remediation under anaerobic conditions while almost negligible under aerobic. Further, these microbes were inoculated in broth containing 1500mg/l nitrate equivalent to the maximum reported nitrate in Yamuna River. E. coli and Enterobacter aerogenes remediated 44% and 46% of 1500mg/l nitrate respectively. Conclusion: These two microbes were found to have moderate potential to remediate nitrate (1500mg/l) which brings novelty to this study. A further detailed study on using microbial consortium to enhance the capability of nitrate remediation will be useful.
Keywords: Nitrate, Fertilizers, Blue Baby Syndrome, Mutation, Bioremediation, Consortium
Full Text:
Introduction
Nitrogen is one of the essential nutrients required by plants for their physiological activity, but its availability is limited.1 Plants uptake nitrogen from the soil in the form of nitrate (NO3-) and ammonium ion (NH4+).2 Therefore, chemical fertilizers (nitrogenous fertilizers) are required to boost the fertility of soil 3 and its sufficient amount needed to grow food for the increasing human population.4The use of chemical fertilizers has increased by 80% from 1960 to 2000.5 This overuse of nitrogenous fertilizers has a hazardous effect on the nitrogen cycle, and this also has degraded water quality around the world.6 Ammonium nitrate, ammonium sulfate and urea are the commonly used nitrogenous fertilizers. Ammonium (NH4+) is a positively charged ion that is quickly absorbed by soil and with the help of soil microflora, converts ammonia into nitrate (nitrification).7 Nitrate (NO3-) is a negatively charged ion and its highly soluble property makes it mobile as it easily mixes with groundwater.8
There are various detrimental effects of nitrate load in drinking water like- Methemoglobinemia in infants commonly known as Blue Baby Syndrome7, Non-Hodgkin Lymphoma,9 respiratory problems, birth defects, tumour in children10, genetic mutation etc. Besides these, the presence of nitrogenous compounds in water leads to eutrophication which is an anaerobic condition responsible for the death of aquatic animals and plants. In wake of these problems, World Health Organization (WHO) set a maximum limit of nitrate in public drinking water as 50mg/l nitrate or 11.3mg/l as nitrate-nitrogen.11The permissible limit of nitrate is 50mg/l for drinking water and no relaxation for the maximum limit. The maximum reported concentration of nitrate in groundwater, Delhi is 1500mg/l.12 Various conventional technologies available to remove nitrate are ion exchange resins, electrodialysis, distillation and reverse osmosis.13 These can remove nitrate, but they produce secondary wastes. Thus, the worthwhile alternative for the denitrification process lies in bioremediation as it is cost-effective and safe.14
Several denitrifying microbes have been reported to remediate nitrates such as E. coli, Enterobacter aerogenes, Shigella sonnei, Klebsiella pneumonia and Enterobacter amnigenusetc. These all belong to the Enterobacteriaceae family which are Gram-negative and rod-shaped(bacilli). According to the reported data mostly nitrate-reducing microbes are bacilli in nature.15E.coli and Enterobacter amnigenus16have the potential to remediate nitrate. The current study aims to isolate and identify such microbes from the Okhla barrage which is one of the most polluted sites of the Yamuna river.17
Materials and Methods
Water quality assessment
The physical parameters were visually analysed while chemical parameters were analysed by using a water testing kit (CPCB water testing kit) to examine the water quality of the Yamuna river. Physical parameters checked were colour, odour, turbidity, and pH, and chemical parameters evaluated were Alkalinity, Hardness, TDS, Chloride, nitrates, phosphates, ammonia, etc.
Chemicals: Potassium Nitrate (96% used for nitrate standard) and sulphuric acid (95%) from Qualigens. Salicylic acid (99%) and Nitrate broth from CDH and Himedia respectively. Nutrient broth from CDH. Pure water used was from the Milli Q Biocel unit (Millipore SAS, Malsheim, France).
Nitrate broth containing Peptone-5g, Beef extract-3g, Potassium nitrate-1g/l, pH(7.0). Nutrient broth containing the following: Peptic digest of animal tissue-5g, Beef extract-1.50g, Yeast extract-1.50g, NaCl-5g/l, pH(7.4).
Collection and isolation of bacterial species
Water samples were collected from the polluted site of Okhla barrage reported as one of the most polluted sites of Yamuna River (CPCB 2017)18 (Table I). Water samples were serially diluted and transferred to nutrient agar containing 500mg/l nitrate and incubated at 37°C for 24hrs. After incubation, bacterial colonies were picked up and then sub-cultured for pure culture isolation.
Standard graph for nitrate
3.6g of KNO3 dried at 105°C for 24hrs. It was then weighed and dissolved in 500ml of deionized water and labelled as a 1000mg/L nitrate-nitrogen solution. Nitrate-nitrogen into nitrate and nitrate into nitrate-nitrogen vice versa calculations were done using the formula:
Nitrate = Nitrate-nitrogen ×4.43
Nitrate-nitrogen= Nitrate ×0.226
Isolation of bacteria and estimating its nitrate remediation potential
Four different isolates showed tolerance up to 500mg/l nitrate concentration. These isolates were further checked for their nitrate remediation potential.19 Each isolate was inoculated (4% inoculum) in Nitrate broth and incubated for 72hrs (3 days) at 120rpm under both aerobic and anaerobic conditions. The culture was centrifuged at 8609RCF (Relative Centrifugal Force) for 10mins.20 40µl supernatants were taken and 200µl salicylic acid (5% salicylic acid in sulphuric acid) was added to it. The mixture was vortexed and incubated in dark for 10mins. The reaction was stopped by the addition of 2ml NaOH (4N NaOH). The absorbance was taken at 420nm after 20 mins of incubation.21 The degree of remediation of nitrate was calculated by using the formula:
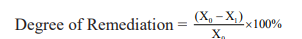
Where X0 is the initial amount of nitrate,
X1 is the amount of nitrate after remediation(72hrs). The final results are calculated with the respect to control samples. (nitrate broth without microorganisms).
To check the remediation rate of isolated microbes whether they are capable to remediate or not 1500mg/l nitrate concentration which is equivalent up to the reported nitrate concentration of Yamuna river.
Phenotypic characterization
These isolates were phenotypically characterized by biochemical tests like Gram stain, colony morphology, nitrate reduction, Indole production, Citrate test, Methyl red test, Voges Proskauer reaction and utilization of carbohydrates (Rita B et al 2009). IMVIC test and eight carbohydrate tests(Glucose, Adonitol, Arabinose, Lactose, Sorbitol, Mannitol, Rhamnose and Sucrose) were analysed using Hi-Media kit (KB001). Metabolic profiling of the microbes helps in identification.
Identification of microbes using 16S rRNA gene sequence
Isolation of Genomic DNA from Bacteria
Each isolated colony was inoculated in 5ml Nitrate Broth and incubated at 37?C at 200rpm in a shaker incubator. 1.5ml bacterial culture was taken in a microcentrifuge tube and centrifuged at 4?C for 10mins at 4000rpm. After centrifugation, the supernatant was removed. Pellet was resuspended in 180µl of lysozyme solution (pH 8.0) followed by incubation at 37?C for 30mins till lysis was observed. Cells were kept at -80?C for 10mins. After that,30µl lysis solution (pH 8.0) was added and incubated in ice for 10mins. An equal volume of Phenol-Chloroform was added (1:1) to separate the DNA from the protein. The tube was centrifuged at 8000rpm for 5mins at 4?C to collect the upper layer.22 Upper layer was taken in another tube and chloroform (500 µl) was added and then again centrifuged. After centrifuging, the upper layer was collected in a new centrifuge tube and 1/10th part of 1M NaCl, 2-2.5 ethanol (absolute ethanol) was added. The tube was kept at -80?C for 30mins and further centrifuged at 10,000rpm for 30mins at 4?C. The supernatant was completely decanted and the DNA pellet was washed with 70% chilled ethanol. Pellet was centrifuged at 8000rpm for 5mins at 4?C then dry pellet and dissolved in 20µl TE buffer.23
Polymerase Chain Reaction
The polymerase chain reaction was used to identify the microbes by isolating bacterial Genomic DNA using Sambrook protocol23,22Universal primer 27F(5`-AGAGTTTGATCCTGGCTCAG-3`) and 1492r (5`- CGGTTACCTTGTTACGACTT-3`) were used in amplification reactions, which were prepared in a total volume of 20µl with the following conditions: -10µl PCR master mix (Genei),0.4µl Primer F, 0.4µl Primer R, 2µl DNA template(50ng/µl) and 7.2µl Nuclease free water (Hyclone) was added. According to thermal profile 95?C 5mins, 95?C 1mins, 58?C 1mins, 72?C 2mins for 30 cycles and followed by 72?C 10mins in Simple Amplifier thermal cycler (Applied Biosystems) and analysed by 1% agarose gel electrophoresis and stained with ethidium bromide under UV light.
Results
Water Quality Assessment
The physical parameters analysed were pH, colour, odour and turbidity (Table II) and the chemical parameters evaluated were Alkalinity, Hardness, chloride, TDS, Fluoride, Iron, Ammonia, Nitrites, Nitrates, Phosphates, Chlorine, BOD and COD. (Table III)
Collection and isolation of bacterial species
The observed concentration of nitrate in water samples collected from the Okhla barrage was found to be between 75-100mg/l.
Eleven different bacterial colonies (PP1-PP11)were identified initially from water samples collected from the Okhla Barrage site on a low concentration of nitrate (range start from 50mg/l). Four, out of these were tolerant to high concentrations of nitrate (up to 500mg/l). They were PP3, 5,7 & 10. The remediation potential of these strains was then checked.
Remediation potential of Bacterial strains
The four bacterial strains which showed tolerance against nitrate were selected to further check their nitrate remediation potential. They were marked as PP3, PP5, PP7 & PP10. 4% inoculum was inoculated in Nitrate broth and incubated at 37°C for 72hrs. After incubation, the optical density of each bacterial strain was measured at 420nm along with control. The nitrate remediation rate of each bacterial strain was checked. PP3, PP5, PP7 and PP10 showed negligible nitrate remediation under aerobic conditions. PP3,PP5,PP7 and PP10 showed remediation of 34.1%, 42.9%, 33.6% and 48.4% respectively under anaerobic conditions (Graph I). Each microbe was then phenotypically characterized.
Further, these four bacterial strains were inoculated in nutrient broth containing 1500mg/l nitrate and then incubated under the same above conditions to check their remediation rate. It was seen that PP5 and PP10 showed 44% and 46% nitrate remediation rates while the rest of the two PP3 and PP7 did show slight remediation potential.
Phenotypic Characterization
All microbes were found to be Gram-negative and rod-shaped bacteria. They belong to the Enterobacteriaceae family based on the results of biochemical tests which are shown in Table IV.
Genotypic characterization of microbes
These isolates were chosen further to identify the genus and species of microbes and also check similarity percentage with strains using BLAST. Identification of isolates was done using phylogenetic analysis of 16S rRNA sequences. Genomic DNA and PCR band of isolates (PP3, PP5, PP7 and PP10) are shown in figure I.
Phylogenetic analysis
The partial 16S rRNA gene sequence of 1399 base pairs of strain PP10 sequence showed 99% similarity with Enterobacter aerogenes (KP764198). 1476 base pair of strain PP3 had 99% similarity with Lelliotiaamnigena and 1409 base pair of PP5 had similarity with E.coli K12. 1398 base pair of PP7 strain had similarity with Klebsiella oxytoca. A Phylogenetic tree was constructed using Clustal W (multiple sequence alignment) and MEGA software. The phylogenetic tree was inferred from 16S rRNA sequences data using neighbour-joining tree are shown in figure II.
Nucleotide Sequence Accession Number
The nucleotide sequence of isolates PP3, PP5, PP7and PP10 were deposited in the GenBank nucleotide sequence database under accession number MN647560, MN754025, MT457847, and MN252552 respectively.
Statistical Analysis
One-way analysis of variance(ANOVA) was used to compare among different groups. Microsoft Excel was used for data presentation. Each experiment was repeated three times in replicates. Mean and standard deviation were calculated and associated probability (P) <0.05 was statistically significant.
Discussion
Isolated microbes belonged to the Enterobacteriaceae family and were found to have the potential to remediate nitrate under anaerobic conditions. Though, they revealed negligible nitrate remediation under aerobic conditions. In the primary screening, eleven strains were isolated from water samples that have tolerance against nitrate. Out of these, four strains showed moderate nitrate remediation potential. On molecular characterization, these strains were identified as E.coliK12, Enterobacter aerogenes, Lelliotiaamnigena and Klebsiella oxytoca. The novelty in this study is that the isolated strains (i.e., E.coli K12 and Enterobacter aerogenes) can remediate 44% and 46% of 1500mg/l (339mg/l nitrate-nitrogen) nitrate concentration. So, the remediation capability on this nitrate concentration showed by isolates have not been reported until now. Further detailed study of these microbes is required to enhance the nitrate remediation potential by addition of some carbon sources and consortiums like bacterial-bacterial association.
Conclusion
This study was a successful trial in isolating and identifying adequate nitrate-eliminating bacteria from polluted sites. E.coli and Enterobacter have shown ~45-50% remediation rate under anaerobic conditions against maximum reported nitrate concentration of Yamuna river (i.e., 1500mg/l). No microbes have been found effective remediation at this concentration, till now. Hence, they can be used in diminishing the nitrate load from groundwater and surface water. Moreover, focus on optimizing the remediation conditions of isolates might lead to great results. Even the association of two or more microbes may impact a lot.
Author’s contribution:-All work regarding the development of methods, optimization, the framework of research preparation, review and editing etc are carried out by the authors.
Acknowledgements
Both authors contributed to the research work and would like to acknowledge the Jaypee Institute of Information Technology for providing financial support. The authors are grateful to the journal’s authors from where the literature of this paper has been reviewed and discussed. The authors are also thankful to IJCRR editorial board members and IJCRR team reviewers who have helped to bring quality to this manuscript.
Source of Funding
Jaypee Institute of Information Technology, Noida
Conflict of Interest
There are no conflicts of interest.
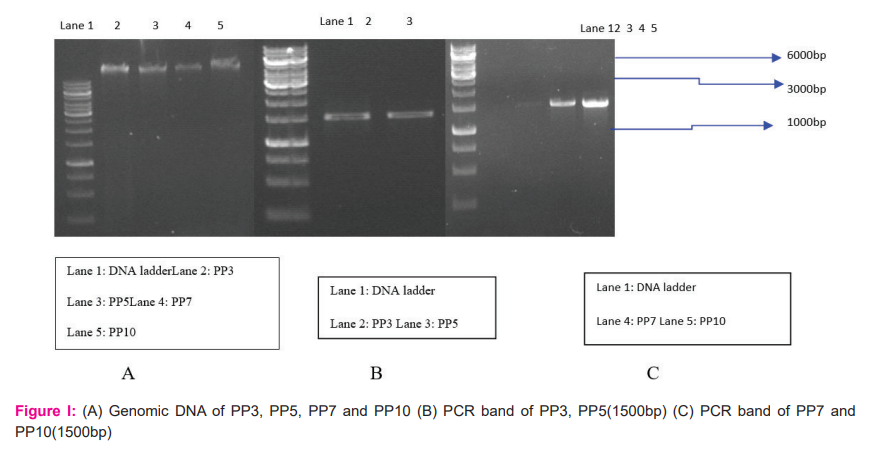
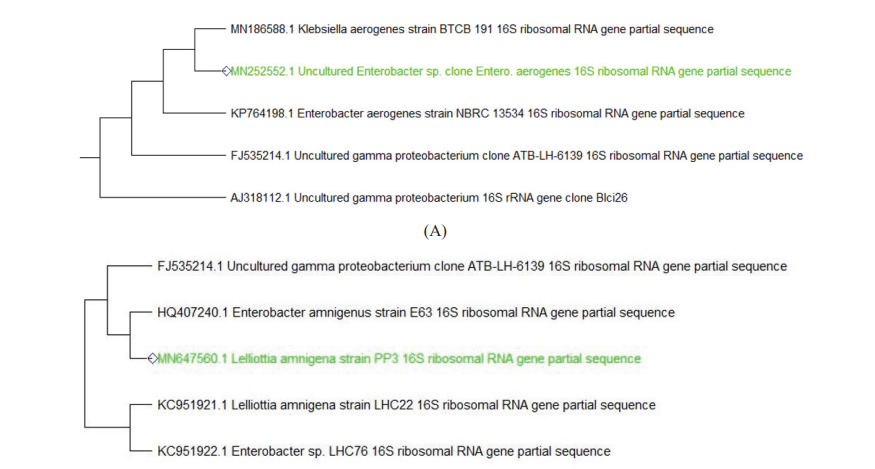
 Figure II: -Neighbor-joining trees showing phylogenetic relationships of 16S rRNA gene sequences to closely related GenBank sequences. These trees were prepared using MEGA software. Green Highlight showing the isolated microbes. (A) Phylogenetic tree of Enterobacteraerogenes (B) Phylogenetic tree of Lelliottiaamnigena (C) Phylogenetic tree of Klebsiella oxytoca (D) Phylogenetic tree of E.coli K12
Figure II: -Neighbor-joining trees showing phylogenetic relationships of 16S rRNA gene sequences to closely related GenBank sequences. These trees were prepared using MEGA software. Green Highlight showing the isolated microbes. (A) Phylogenetic tree of Enterobacteraerogenes (B) Phylogenetic tree of Lelliottiaamnigena (C) Phylogenetic tree of Klebsiella oxytoca (D) Phylogenetic tree of E.coli K12
References:
1. Sharma S, Jha PK, Ranjan MR, Singh UK, Jindal T, Sakram G, et al. Nitrate removal efficiency of bacterial consortium (Pseudomonas sp. KW1 and Bacillus sp. YW4) in synthetic nitrate-rich water. Bioresour. Technol. [Internet]. 2017;157(2):553–63. Available from: http://dx.doi.org/10.1016/B978-0-12-818307-6.00010-X
2. Chadwick DR, John F, Pain BF, Chambers BJ, Williams J. Plant uptake of nitrogen from the organic nitrogen fraction of animal manures: A laboratory experiment. J Agric. Sci. 2000;134(2):159–68.
3. Savci S. An Agricultural Pollutant?: Chemical Fertilizer. Int. J. Environ. Sci. Technol 2012;3(1):11–4.
4. Galloway JN, Townsend AR, Erisman JW, Bekunda M, Cai Z, Freney JR, et al. Transformation of the nitrogen cycle: Recent trends, questions, and potential solutions. Science (80- ). 2008;320(5878):889–92.
5. Brender JD, Weyer PJ, Romitti PA, Mohanty BP, Shinde MU, Vuong AM, et al. Prenatal nitrate intake from drinking water and selected birth defects in offspring of participants in the national birth defects prevention study. EnvironHealth Perspect. 2013;121(9):1083–9.
6. Kim HR, Yu S, Oh J, Kim KH, Lee JH, Moniruzzaman M, et al. Nitrate contamination and subsequent hydrogeochemical processes of shallow groundwater in agro-livestock farming districts in South Korea. Agric. Ecosyst Environ [Internet]. 2019;2720:50–61.Available from: https://doi.org/10.1016/j.agee.2018.12.010
7. The D, Parameters C, Quality OF, Drinking OF, In ACS. OF QUALITY OF DRINKING WATER?; A CASE STUDY in Int. J Curr Res Rev. 2011;03(09):33–6.
8. Lin B Le, Sakoda A, Shibasaki R, Suzuki M. A modelling approach to global nitrate leaching caused by anthropogenic fertilisation. Water Res. 2001;35(8):1961–8.
9. Majumdar D, Gupta N. Nitrate pollution of groundwater and associated human health disorders. Indian J Environ. Health. 2000;42(1):28–39.
10. Knekt, P, Järvinen R, Dich J, Hakulinen T. Risk of colorectal and other gastro?intestinal cancers after exposure to nitrate, nitrite and N?nitroso compounds: a follow?up study. Int J Cancer. 1999;80(6):852–6.
11. Isibor RA, Aderogbin JA. Assessment of Nitrate and Escherichia Coli Contamination of Shallow Groundwater of Ajakanga and Environs, Ibadan, Southwestern Nigeria. Iconic Res Engg. 2020;3(11):108–18.
12. Kumar L. A study of nitrate contamination in groundwater of Delhi, India. Vol. 10, Asian J Water Envt Poll. 2013;91–4.
13. Ward MH, Jones RR, Brender JD, de Kok TM, Weyer PJ, Nolan BT, et al. Drinking water nitrate and human health: An updated review. Int. J. Environ. Res. Public Health. 2018;15(7):1–31.
14. DebRoy S, Das S, Ghosh S, Banerjee S, Chatterjee D, Bhattacharjee A, et al. Isolation of nitrate and phosphate removing bacteria from various environmental sites. Online J. Biol. Sci. 2012;12(2):62–71.
15. Lin JT, Stewart V. Nitrate assimilation by bacteria. Adv Microb Physiol. 1997;39:1–30.
16. Fazzolari E, Mariotti A, Germon JC. Nitrate reduction to ammonia: A dissimilatory process in Enterobacter amnigenus. Can J Microbiol. 1990;36(11):779–85.
17. Sarkar A, Ali S, Kumar S, Shekhar S, Rao SVN. Groundwater Environment in Delhi, India. Groundwater Environment in Asian Cities: Concepts, Methods and Case Studies. Elsevier Inc.; 2016. 77–108 p.
18. Sharma S, Jha PK, Ranjan MR, Singh UK, Jindal T. Water Quality Monitoring of Yamuna River by Using GIS-Based Water Quality Index in Delhi, India. Int J Curr Microbiol Appl Sci. 2017;6(2):1249–63.
19. F.A.J. Armstrong. Determination of nitrate in water ultraviolet spectrophotometry. Anal Chem. 1963;35:1292–4.
20. Strong PJ, Burgess JE. Treatment methods for wine-related and distillery wastewaters: A review. Bio Remediat J. 2008;12(2):70–87.
21. Chouhan S, Tuteja U, Flora SJ. Isolation, identification and characterization of fluoride resistant bacteria: possible role in bioremediation. Appl Biochem. Microbiol. 2012;48(1):51–8.
22. Pitcher DG, Saunders NA, Owen RJ. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett Appl Microbiol. 1989;8(4):151–6.
23. Code C. A Practical Approach to Molecular Biology Course Code?: 07B41BT708.
|






 This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License