IJCRR - 13(11), June, 2021
Pages: 65-75
Date of Publication: 04-Jun-2021
Print Article
Download XML Download PDF
Mining Plausible Antibacterial Targets Against Potato Pathogen Ralstonia solanacearum IPO1609 Through in Silico Subtractive Genomics Approach
Author: Gurunathan S, Dhamotharan R
Category: Healthcare
Abstract:Introduction: Ralstonia solanacearum IPO1609 (Rs IPO1609) is a gram-negative phytopathogenic bacteria that causes severe bacterial wilt disease in potatoes. Agricultural practices and agrochemicals are often ineffective solutions to control it. Identification of essential antibacterial targets in the phytopathogen could enable the design/development of suitable bactericides and eventually, control potato brown rot. Objective: To reveal prospective antibacterial targets in Rs IPO1609 utilizing subtractive genomics strategy coupled with differential pathway analysis, subcellular localization, virulent prediction and drug bank database screening. Methods: The study was designed to identify potential antibacterial targets in Rs IPO1609. Among the 4545 proteins present
in the pathogen, non-orthologs cum non-paralogs were obtained and subjected to in silico comparative analysis against potato
proteome to reveal non-homologous proteins present in the bacterium. Furthermore, the essentiality of these non-homologs for
pathogen's survival was determined using DEG Database. Metabolic pathways involvement of the short-listed essential proteins
was implemented using KAAS and virulent proteins were determined using MP3 web server analysis. Intracellular localization of
essential virulent proteins determined using CELLO2GO & PSORTb programs enable enlisting of plausible antibacterial targets.
Results: Subtractive genomics-based approach revealed that a list of 136 proteins of Rs IPO1609 were potato non-homologs
and essential. A total of 55 targets are involved in the unique biochemical pathways of the pathogen. Of 55, 29 proteins were found virulent. Furthermore, based on intracellular localization, 3 virulent proteins were identified as promising therapeutic targets. Conclusion: Among the 55 targets identified in this study, three proteins were found highly potential as antibacterial targets in
Rs IPO1609 based on their metabolic pathway, virulence and intracellular localization properties. The outcome might be used as
a design in genomics-based strategies to control bacterial phytopathogens.
Keywords: Ralstonia solanacearum IPO1609, Antimicrobial targets, Subtractive genomics, Essential genes, Potato pathogen, KEGG pathway
Full Text:
INTRODUCTION
Current strategies to manage bacterial plant diseases are becoming obsolete primarily due to deficiency in availability of suitable agrichemicals,1 resulting in an overall loss of twenty per cent (approx.) in plant productivity globally.2 Thus, forcing researchers to explore and identify novel agrichemicals to control bacterial phytopathogens. In today’s era, computational approaches have massively supported modern medicinal drug discovery and development processes.3 However, investigators express that in silico strategies are vital in exploring and understanding plant sciences too.4 Few even predict applying computational protocols to decode phytopathogens, their molecular virulence factors and discover effective therapeutics to control them.5,6
Antimicrobial target discovery in today’s post-genomics era is achieved by ‘omics’ based approaches rather than by the traditional generic methods.7 This strategy is also being explored against phytopathogens.1 Genomic data of several bacterial phytopathogens (draft/completed) generated by cutting-edge sequencing technologies are publicly accessible.1,2 Thus, allowing investigators to implement computational strategies (especially in silico comparative, subtractive, and functional genomics) on these collected data to discover novel antibacterial targets.8–11
Promising antimicrobial targets in a pathogen can be detected with an interesting approach known as ‘Differential genome display’ or ‘Subtractive Genomics’.12 This approach filters gene(s) (or its protein products) essentially needed for the pathogen, but absent in the host organism (also termed as essential non-host homologous sequences) and thus, are regarded as worthy targets against the pathogen.13 Although experimental and computational strategies exists for essential gene-based target prediction, the latter is preferred as it involves less time, labour and economic.14,15 Computational target identification in human bacterial pathogens has been successfully implemented by several investigators.16–23 However, only a few researchers have followed this strategy to recognize antibacterial targets in phytopathogens.8–10
Ralstonia solanacearum (Rs) is a heterogeneous group of bacterial phytopathogen causing the most devastating wilt disease globally in more than 450 economically important plants that include banana, tomato, potato, eggplant, groundnut and tobacco. This soil bacterium is gram-negative, aerobic, non-spore-forming motile bacilli and belongs to the ?-proteobacteria family. More than 140 Rs strains recognized worldwide are classified into five races (1, 2, 3, 4 and 5), six biovars (1, 2, 3, 4, 5 and 6) and four phylotypes (I, II, III and IV) based on their ability to infect different hosts, biochemical properties and geographical distribution,24–26 respectively. The phylotypes are further subgrouped into different sequevars.24,25,27
Rs IPO1609 strain is a race 3/biovar 2 isolate obtained from potato in Europe and has been recently classified as phylotype IIB sequevar 1 (IIB1) strain. In addition to being highly destructive among known Rs strains affecting potato and ability to adapt to highland temperatures, IIB1 are reported as highly dangerous potato pathogens because they cause asymptomatic latent infections.28 A whole-genome sequence draft (20x) of Rs IPO1609 was established at Genoscope, France.29 During the sequencing process, only the final assembling step was not completed. Hence, these sequences were assembled to generate ten supercontigs (length of 4 to 3372 kb) and are deposited in NCBI GenBank (accession nos.: CU694431 - CU694438, CU914166 and CU914168). They are also accessible at iant.toulouse.inra.fr/bacteria/annotation/cgi/ralso.cgi. Over 99% of the 5.313 Mb genome sequence is covered by 6 supercontigs (each having >10 kb length) alone. The genome seems to possess a high G+C content (average 60%).29
The present work aims to utilize a subtractive genomics strategy on Ralstonia solanacearum IPO1609 (a potato brown rot causing pathogen) to reveal its prospective antimicrobial target candidates. Furthermore, employing differential pathway analysis, subcellular localization, virulent prediction and drug bank database screening on the plausible targets will identify novel therapeutic targets among them. Eventually, facilitating the discovery of suitable therapeutics to use against this important bacterial phytopathogen.
MATERIALS AND METHODS
Data Collection
All protein sequences of Ralstonia solanacearum IPO1609 were retrieved individually in Fasta format from http://iant.toulouse.inra.fr/bacteria/annotation/cgi/ralso.cgi. The whole Proteome of Solanum tuberosum (UP000011115) was obtained from UniProtKB database (http://www.uniprot.org/taxonomy/complete-proteomes). All essential genes associated with prokaryotes were downloaded from the Database of Essential Genes (DEG) version 15.2 (http://tubic.tju.edu.cn/deg/).30
Mining Essential Proteins of Rs IPO1609
Exclusion of Paralogs and Orthologs
CD HIT server analysis (http://weizhong-lab.ucsd.edu/cdhit_suite/cgi-bin/index.cgi?cmd=cd-hit)31 at 80% identity threshold21 was employed to screen duplicate proteins or paralogs present in the proteome of Rs IPO1609. Likewise, orthologs (i.e. protein sequences with <=100 amino acids) among the protein sequences of Rs IPO1609 were also examined.16,22,23 These identified paralogs and orthologs were excluded from Rs IPO1609 proteome dataset.
Screening of Non-Host Homologs
All the non-paralog cum non-ortholog proteins of Rs IPO1609 resulting from the previous step were analysed using BLASTP tool32 to detect their similarity with the host i.e. potato (Solanum tuberosum) proteins. A 0.01 random e-value (expectation value) threshold was fixed. Pathogen proteins showing ‘no hits found’ during BLAST analysis form the resultant dataset and are termed as ‘Non-Host Homologs’.
Extraction of Non-Host Homolog Essential Proteins
Extraction of essential proteins from non-host homologs of Rs IPO1609 was achieved by BLAST search (e-value <= (1/1010)9 and 30% or more sequence identity22) against all prokaryotic sequences of the DEG database. The resulting protein hits from Non-Host Homolog Essential Proteins of Rs IPO1609.
All BLAST searches utilized Stand-alone BLAST (version 2.6.0+) and the necessary parameters such as e-value, sequence identity, etc. was provided using command-line options. An in-house developed PERL script was employed to parse protein sequences from the respective BLAST results.
Metabolic Pathway Studies
Each Non-Host Homolog Essential Protein of Rs IPO1609 was submitted as a query sequence to KAAS (KEGG Automatic Annotation Server) at KEGG (http://www.genome.jp/tools/kaas). Functional annotation of each query protein is identified by searching against KEGG GENES database and the results are reported as KEGG Orthology (KO) assignments and KEGG Pathways.33 Unique pathways of Rs IPO1609 are revealed by manual comparison of pathogen and potato metabolic pathways listed in KAAS results. As the remaining pathways are part of only host cells or common to both (host and pathogen), hence, can be considered insignificant. So, they are not taken up further in our study. Thus, essential proteins associated with unique pathways of the pathogen alone were enlisted as possible therapeutic targets.
Prediction of Virulent Proteins
The disease-causing ability of a microbe is dependent on its virulent factors, especially proteins produced by them. Thus, predicting the virulence nature of a protein will enable us to classify virulent ones as better therapeutic targets. So, each putative therapeutic target protein identified was subjected to MP3 web server analysis.34 This online tool integrates Support Vector Machine and Hidden Markov Model approach for fast, accurate and sensitive prediction of bacterial virulent proteins.
Subcellular Localization Prediction
Intracellular localization of target proteins in the pathogen was predicted using PSORTb tool (version 3.0)35 and CELLO2GO server (http://cello.life.nctu.edu.tw/cello2go).36 results from both the web tools have higher accuracy. Thus, each protein in the dataset was segregated based on its subcellular localization.
The overall workflow involved in genomic subtraction based antibacterial target identification in Ralstonia solanacearum IPO1609 is depicted in Figure 1.
RESULTS
Identification of Non-Host Homolog Essential Proteins in Rs IPO1609
A total of 4545 proteins encoded by Rs IPO1609 genome were individually retrieved. 322 orthologs and 127 paralogs of the pathogen were identified and excluded. Thus, 9.87% of the pathogen’s proteins were removed from the dataset. The remaining 4096 proteins of the bacterium BLASTed against 53,105 proteins of the potato (host) proteome reveals 2530 ‘Non-Host Homologous Proteins’ of the pathogen. Thus, 34.4% of bacterial proteins homologous to the host were eliminated from further analysis. BLAST Alignment of Non-Host Homologs against DEG’s prokaryotic protein sequences (18,835 sequences) recognizes 136 essential proteins of the pathogen (2.9%) as Non-Host Homolog Essential Proteins vital for Rs IPO1609 survival. Table 1 displays results obtained during different stages of subtractive and metabolic pathway analyses performed in this study.
Unique Metabolic Pathway(s) Involvement of Rs IPO1609 Essential Proteins
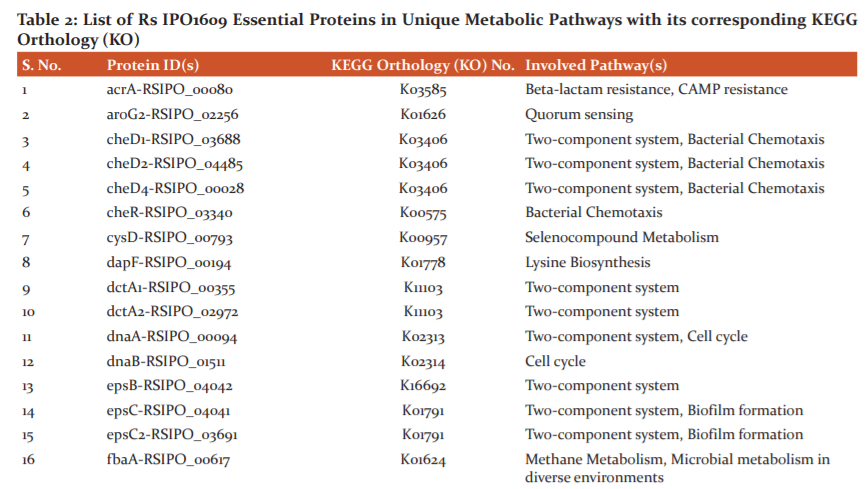
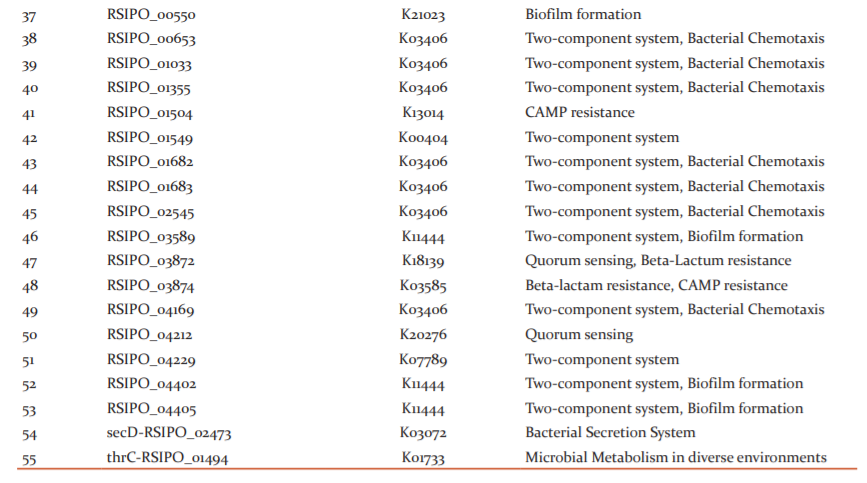
The role of shortlisted essential proteins in different biochemical pathways were determined using the KAAS server. Among 136 proteins analyzed, 126 were involved in different biochemical pathways of the pathogen. Several proteins were involved in primary metabolic pathways such as oxidative phosphorylation, amino acid biosynthesis, citric acid cycle, etc. Some proteins were associated with peptidoglycan biosynthesis, LPS biosynthesis, bacterial chemotaxis, flagellar assembly, protein export and secretion systems. A total of 16, 13 and 7 proteins were identified to be involved in the two-component system, biosynthesis of secondary metabolites and ABC transporters mechanisms, respectively. However, KO details of ten essential proteins couldn’t be established and so, were excluded from further analysis.
The involvement of each chosen essential protein in unique metabolic pathways of Rs IPO1609 was also determined. A total of 59 biochemical pathways were identified in the pathogen. Of 59 pathways, 20 (i.e. 33.8%) were characterized as unique. Similarly, among 136 essential proteins, 55 were found participating in several unique pathways of the bacterium. Surprisingly, among 55, 22 essential proteins reveal their role in only one pathway of the pathogen. Whereas, remaining 33 proteins were associated with more than one unique pathway associated with the pathogen (Table 2). Because of their commonness to both pathogen and host, the remaining 39 pathways were not considered for further investigation.
Virulence and Subcellular Localization Prediction Analyses
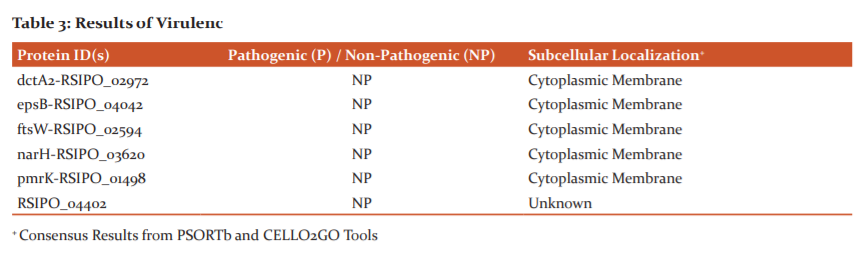
The virulent/Avirulent property of the 55 essential proteins selected was predicted with an MP3 webserver. Our results reveal 29 virulent proteins among the 55 proteins studied (Table 3).
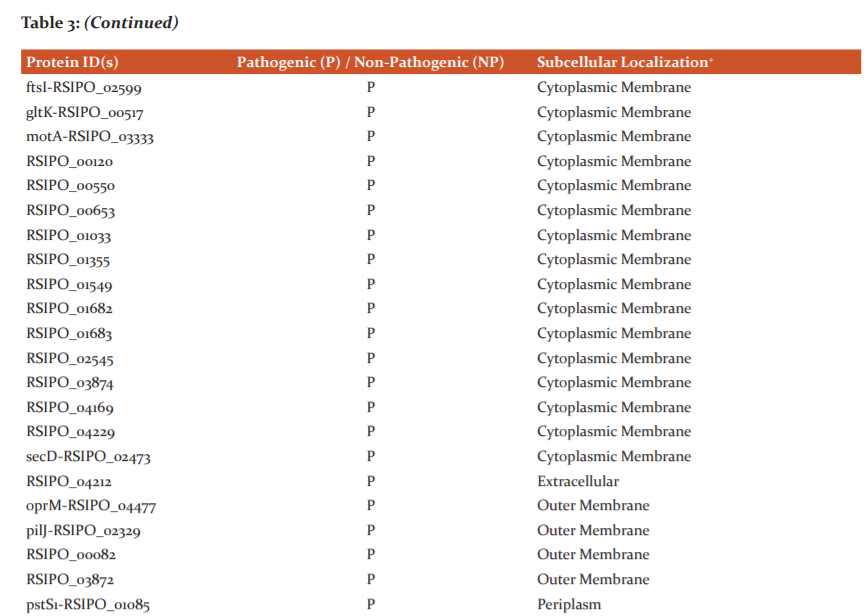
Figure 2 displays the distribution of Rs IPO1609 essential proteins at various subcellular localizations. Table 3 provides consensus results of subcellular localization prediction obtained from PSORTb and CELLO2GO programs. Results reveal 49% of query proteins localize in the cytoplasmic membrane and 36% localize in the cytoplasm. Only 7% of proteins were found to localize in the outer membrane region of the cell. An equal number of proteins (i.e. 2%) was predicted as molecules associated with extracellular and periplasm zones. However, the subcellular localization of 4% proteins could not be determined.
Analyzing results of virulence and subcellular localization prediction renders only three proteins viz. one extracellular (Id: RSIPO_04212) and two cytoplasms (Id: gspE-RSIPO_02864, RSIPO_04405) localizing as highly plausible therapeutic targets among the studied 55 essential proteins.
DISCUSSION
In the drug discovery phenomenon, antimicrobial target identification is a primary step that is both sensitive and critical.37 In this post-genomics era, investigating “omics” data combined with advanced computational protocols has enabled easier and improved identification of protein targets in several pathogens.16,38,39 One such popular approach referred to as ‘subtractive genomics’ has been employed in effective target predictions in human pathogens.16–23,40 Over the last couple of decades, ‘omics’ data on bacterial phytopathogens obtained from various sequencing projects has generated renewed interest to apply in silico research in identifying antibacterial targets.41,42 Nevertheless, only a few investigators have utilized in silico essential-gene based target identification in phytopathogenic bacteria.8–10 To the best of our knowledge, no literature is available on the computational identification of novel antimicrobial targets in potato brown rot causing bacterial pathogen Rs IPO1609. Thus, the current study applies a subtractive genomics strategy on the Rs IPO1609 proteome to reveal its’ plausible targets.
The suitability of antimicrobial targets identified by genome subtraction technique is weighed upon two fundamental criteria viz. ‘Selectivity’ and ‘Essentiality’. Any protein exclusively present in a pathogen as well as is necessary for its basic survival makes an effective antimicrobial target.43 Hence, an initial step in this investigation focused on identifying proteins that are both specific and essential to Rs IPO1609 strain. Among the 4545 proteins of the pathogen, 4096 non-paralogs cum non-orthologs were identified. Since paralogs are redundant as antimicrobial targets21 and orthologs have little chances of being essential to the pathogen,16,22,23 both were eliminated from the study. A total of 2530 proteins of the pathogen were identified as ‘Non-Host Homologs’ following BLAST analysis against potato host proteome. Previously, only 158 proteins in Pseudomonas syringe pv phaseolicola were recognized as non-homologs.8 Similarly, only 406 and 152 sequences were non-homologs in two different phytopathogenic strains of Xanthomonas oryzae studied.9,10 Several earlier literature report between a few hundred and a few thousand proteins identified as non-homologs in several gram-negative bacterial members.19–23,38,44
According to Luo and coworkers,30 essential proteins of microbes are regarded as interesting antimicrobial targets. Thus, 136 essential proteins among the ‘Non-Host Homologs’ of the pathgen were identified by BLAST Alignment against the DEG database. In 2004, 300 – 400 genes were identified as essential in Pseudomonas aeruginosa.13 Whereas, only 137 essential proteins were reported in Pseudomonas syringae pv phaseolicola.8 However, less than 40 proteins were found essential in two different strains of Xanthomonas oryzae.9,10 Several researchers document varying essential protein numbers identified by them in gram-negative bacteria.8–10,18,44 Number of Non-Homologs, as well as essential proteins identified by different investigators, seems to vary due to the differences in the computational protocols followed as well as experimental parameters employed by them.
According to Barh and coworkers,22 Essential Non-Host Homologs identified during in silico subtractive/comparative genomics analysis has been the major criterion in establishing therapeutic targets. However, advancements in insilico resources of the preceding decade have facilitated researchers to opt for additional factors in determining the suitability of antimicrobial targets.38,40,45 Thus, essential proteins that are associated with pathogen-specific biochemical pathways, their subcellular localization, virulence and druggable properties were employed as additional factors by previous investigators. Hence, these characters for all the enlisted essential proteins were determined.
KAAS analyses of the 136 essential proteins reveal the involvement of 126 proteins in different biochemical pathways of the pathogen; many engaged in primary metabolic pathways and few in other pathways. These processes have a direct or indirect link to motility, virulence factor, pathogenesis and nutrient mobilization/uptake mechanisms of the bacterium. 46,47
Essential proteins engaged in unique metabolic pathways of Rs IPO1609 were determined, as they are considered good antibacterial targets. A total of 55 essential proteins participate as metabolites in 20 unique metabolic pathways of the pathogen. Since these molecules are part of pathways indispensable for bacterial life, they form interesting targets. Less than 50 essential proteins have been reported to be involved in exclusive pathways of several gram-negative bacteria.16,20,38 In phytopathogen Pseudomonas syringae pv phaseolicola, 22 essential proteins with a role in pivotal metabolic pathways were documented.8 Analogous to earlier reports, several essential proteins associated with pathways of both i.e. host and pathogen (called common host-pathogen pathways) were also revealed.16,17,22,38 These proteins, if chosen as targets, may also damage host cells, so, were eliminated from the dataset.
A protein of the pathogen can find an active role in single or multiple pathways and more than one protein can involve in one or several metabolic pathways. Results in our investigation revealed 22 proteins participating in only one pathway. However, the remaining 33 proteins were associated with multiple pathways of the pathogen and can be regarded as significant antimicrobial targets.22
A two-component system in a bacterium is a signal transduction machinery that mounts a suitable response identifying changes in a cell’s exterior or interior.48 7 Rs IPO1609 proteins associated with a two-component system were identified and may be regarded as superior antibacterial targets as they are essential for bacterial survival. Similarly, 12 chemotaxis proteins necessary to sense chemical gradients in their environment and facilitate movement towards favourable conditions were recognized as potential targets in Rs IPO1609. Additionally, PhosphoTransferase System (PTS) and Bacterial Secretion System in Rs IPO1609 are among its identified unique metabolic pathways. As these pathways are pivotal for the growth and survival of a bacterium in extreme conditions, they form significant antibacterial targets. Our results are in concurrence with earlier reports.16,20,22,49
Peptidoglycan in the cell wall of a bacterium helps the cell to maintain its structure as well as resists osmosis. Being an important virulence factor of the bacterium, compounds inhibiting the proteins participating in peptidoglycan biosynthesis can be effective antimicrobials.16,17,22,38 Our results prioritize two unique proteins associated with Rs IPO1609 peptidoglycan biosynthesis pathways and might be recognized as plausible targets.
In structural terms, LipoPolySaccharides (LPS) consists of a core oligosaccharide linked to Lipid A and O-antigen molecules on either ends and is responsible for gram-negative bacteria outer membrane stability.50 Our results list three proteins as key metabolites in the LPS biosynthesis pathway of Rs IPO1609. Thus, these proteins may have opted as significant targets for therapeutics discovery. Antimicrobials blocking the permeability of solutes across the membrane results in cell death. Results obtained in our study are in parallel to earlier literature reports.16,20,22,38
Autoinducers help communication among bacterial cells and their concentration is directly proportional to bacterial proliferation. Once a threshold is reached, increased virulence occurs resulting in biofilm formation.51 Chemicals targeting quorum sensing (QS) prevents in vivo QS activation, eventually leading to a decrease in virulence. Since bacterial growth is not directly controlled by QS, blocking them does not compel the emergence of antimicrobial resistance in target organisms.52,53 Achieving QS inhibition (aka Quorum quenching) by blocking receptors using antagonistic molecules, blocking autoinducer synthesis and degradation of autoinducers with hydrolytic enzymes, eventually affects the formation of biofilms.53 In this study, five proteins of Rs IPO1609 recognized as metabolites of the QS mechanism might be regarded as significant antimicrobial targets. Recently, Pseudomonas aeruginosa QS proteins were suggested as attractive antibacterial targets.18
Cationic antimicrobial peptide (CAMP) resistance and beta-lactam resistance-conferring proteins occurring in Rs IPO1609 suggest that the pathogen possesses a resistance mechanism towards antimicrobial molecules. Despite CAMP’s ability to weaken bacterial cell membrane integrity, many bacteria have evolved alternative pathways to resist CAMP molecules54. In this computational study, four CAMP resistant proteins with potentiality as therapeutic targets have been identified. Recently, Prabha and team10 reported CAMP as possible targets in a rice pathogen. Beta-lactam proteins are considered as antibiotics targeting cross-linkage in the peptidoglycan layer of a bacterial cell wall. Thus, beta-lactam resistance conferred by proteins elaborated by a bacterium is a vital virulence factor. Proteins of these pathways can form as good targets for influencing resistance and thus, rendering the bacterium susceptible. Three proteins responsible for beta-lactam resistance in Rs IPO1609 represent plausible targets in the pathogen. Few other researchers have recorded similar results in bacterial pathogen of humans38 and plants.10
All the 55 essential proteins short-listed through the current study can be regarded as novel potential targets. However, the virulent nature, as well as intracellular localization of these proteins, might provide significant details for the drug design and development process. Our results identified 29 virulent proteins among the 55 plausible protein targets. Earlier, Keshri and co-workers9 reported three virulent proteins in Xanthomonas oryzae pv oryzae PXO99A. Similarly, four proteins were found virulent in Xanthomonas oryzae pv oryzae KACC10331 in a recent investigation.10
Extracellular space, outer membrane, periplasmic, cytoplasmic (inner) membrane and cytoplasm are the five probable localizing zones in gram-negative bacteria.36 Our analysis revealed 25 and 2 virulent proteins localizing in membranous and cytoplasm region of the bacterium respectively. However, 1 virulent protein was predicted secreting extracellularly and 1 virulent protein’s localization could not be determined. According to Barh et al.,22 bacterial proteins secreted extracellularly or localizing in the cytoplasm are favourable targets. Thus, three virulent proteins viz. RSIPO_04212 (extracellular), gspE-RSIPO_02864 and RSIPO_04405 (cytoplasm) are plausible antibacterial targets identified in Rs IPO1609.RSIPO_02864 (gspE) is a well-known protein involved in type 2 secretion systems of a bacterium and RSIPO_04405 has been computationally predicted as a signal transduction protein. However, RSIPO_04212 is computationally predicted to be a haemagglutinin/hemolysin related protein. In 2012, Katara and co-workers8 have reported intracellular localization of certain proteins in Pseudomonas syringae. But, their reports lack details on either the proteins involved or their intracellular localization. No other literature regarding localization prediction of bacterial phytopathogen proteins using computational approaches is currently documented. Nevertheless, investigators utilize in silico strategies to predict the localization of proteins in numerous gram-negative human bacterial pathogens.16,17,19,20,22,38 For obtaining more reliable results, researchers have opted for two or more tools during their computational prediction analysis.
CONCLUSION
The subtractive genomics strategy attempted in this research work can act as a preparatory step in the discovery of suitable agrichemicals against the potato pathogen Rs IPO1609. Since the 55 proteins (Table 2) are essential and unique to Rs IPO1609 strain, they can be considered as significant antibacterial targets. Based on virulence and intracellular localization properties, three proteins viz. gspE-RSIPO_02864, RSIPO_04405 and RSIPO_04212 are reported as promising targets. Bactericides targeting these proteins can help to chemically control wilt disease of potato caused by Rs IPO1609. However, validating these targets through experimental studies is required. Our methodology and the results generated can be considered significant to the researchers actively involved in the bacterial genomics-based study for control of phytopathogens.
ACKNOWLEDGEMENTS
The author Gurunathan, S expresses his sincere thanks to the Management of National College (Autonomous), Tiruchirapalli for their constant support and encouragement during this research study. Authors thank the University Grants Commission (UGC), India for the financial support provided in the form of the UGC Minor Research Project (MRP-6785/16). The authors acknowledge the immense help received from the scholars whose articles are cited and included in references of this manuscript.
CONFLICTS OF INTEREST
None
SOURCE OF FUNDING
This research work is supported by a financial grant from the University Grants Commission (UGC), India in the form of UGC Minor Research Project (MRP-6785/16).
AUTHOR’S CONTRIBUTION
SG conceived, designed and performed the experiments. Both the authors analyzed the results, provided critical feedback, contributed to discussion and in shaping the final manuscript.




References:
1. Sundin GW, Wang N, Charkowski AO, Castiblanco LF, Jia H, Zhao Y. Perspectives on the Transition From Bacterial Phytopathogen Genomics Studies to Applications Enhancing Disease Management: From Promise to Practice. Phytopathology. 2016 Oct;106(10):1071–82.
2. Tiwari S, Awasthi M, Pandey VP, Dwivedi UN. Genomics Based Approaches towards Management of Plant Diseases with Emphasis on in silico Methods as a Prudent Approach. J Agri Sci Food Technol. 2017 May;3(3):39–51.
3. Taylor D. The Pharmaceutical Industry and the Future of Drug Development. In: Hester RE, Harrison RM, editors. Issues in Environmental Science and Technology [Internet]. Cambridge: Royal Society of Chemistry; 2015 [cited 2020 Jun 26]. p. 1–33. Available from: http://ebook.rsc.org/?DOI=10.1039/9781782622345-00001
4. Rhee SY, Dickerson J, Xu D. Bioinformatics and its Applications in Plant Biology. Annu Rev Plant Biol. 2006;57(1):335–60.
5. Shanmugam G, Jeon J. Computer-Aided Drug Discovery in Plant Pathology. Plant Pathol J. 2017;33(6):529–42.
6. Imam J, Singh PK, Shukla P. Plant Microbe Interactions in Post Genomic Era: Perspectives and Applications. Front Microbiol. 2016;7:1488.
7. Lin J, Qian J. Systems biology approach to integrative comparative genomics. Expert Rev Proteom. 2007;4:107–19.
8. Katara P, Grover A, Sharma V. In Silico Prediction of Drug Targets in Phytopathogenic Pseudomonas syringae pv. phaseolicola: Charting a Course for Agrigenomics Translation Research. OMICS J Integr Bio. 2012 Dec;16(12):700–6.
9. Keshri V, Singh DP, Prabha R, Rai A, Sharma AK. Genome subtraction for the identification of potential antimicrobial targets in Xanthomonas oryzae pv. oryzae PXO99A pathogenic to rice. Biotechnology. 2014;4(1):91–5.
10. Prabha R, Singh DP, Ahmad K, Kumar SPJ, Kumar P. Subtractive genomics approach for identification of putative antimicrobial targets in Xanthomonas oryzae pv. oryzae KACC10331. Arch Phytopathol Plant Protect. 2019;52(7–8):863–72.
11. Pucci MJ. Use of genomics to select antibacterial targets. Biochem Pharmacol. 2006;71(7):1066–72.
12. Huynen M, Dandekar T, Bork P. Differential genome analysis applied to the species-specific features of Helicobacter pylori. FEBS Lett. 1998;426(1):1–5.
13. Sakharkar KR, Sakharkar MK, Chow VTK. A novel genomics approach for the identification of drug targets in pathogens, with special reference to Pseudomonas aeruginosa. In Silico Biol (Gedrukt). 2004;4(3):355–60.
14. Itaya M. An estimation of minimal genome size required for life. FEBS Letters. 1995;362(3):257–60.
15. Kobayashi K, Ehrlich SD, Albertini A, Amati G, Andersen KK, Arnaud M, et al. Essential Bacillus subtilis genes. Proc Natl Acad Sci USA. 2003;100(8):4678.
16. Hossain MU, Khan M, Hashem A, Islam MdM, Morshed MN, Keya CA, et al. Finding Potential Therapeutic Targets against Shigella flexneri through Proteome Exploration. Front Microbiol. 2016;7:1817.
17. Shuvo MSR, Shakil SK, Ahmed F. Potential Drug Target Identification of Legionella pneumophila by Subtractive Genome Analysis: An In Silico Approach. Bangla J Microbiol. 2019;35(2):102–7.
18. Uddin R, Jamil F. Prioritization of potential drug targets against P. aeruginosa by core proteomic analysis using computational subtractive genomics and Protein-Protein interaction network. Comput Biol Chem 2018;74:115–22.
19. Sadhasivam A, Vetrivel U. Genome-wide codon usage profiling of ocular infective Chlamydia trachomatis serovars and drug target identification. J Biomol Struc Dynam. 2018;36(8):1979–2003.
20. Mondal SI, Ferdous S, Akter A, Mahmud Z, Karim N, Islam MdM, et al. Identification of potential drug targets by subtractive genome analysis of Escherichia coli O157:H7: an in silico approach. AABC. 2015:49.
21. Shoukat K, Rasheed N, Sajid M. Subtractive genome analysis for in silico identification and characterization of novel drug targets in C. Trachomatis STRAIN D/UW-3/Cx. 2012;4.
22. Barh D, Tiwari S, Jain N, Ali A, Santos AR, Misra AN, et al. In silico subtractive genomics for target identification in human bacterial pathogens. Drug Dev Res. 2011;72(2):162–77.
23. Vetrivel U, Subramanian G, Dorairaj S. A novel in silico approach to identify potential therapeutic targets in human bacterial pathogens. HUGO J. 2011;5(1–4):25–34.
24. Peeters N, Guidot A, Vailleau F, Valls M. Ralstonia solanacearum , a widespread bacterial plant pathogen in the post-genomic era: Ralstonia solanacearum and bacterial wilt disease. Mol Plant Pathol. 2013;14(7):651–62.
25. Elphinstone JG. The current bacterial wilt situation: a global overview. In: Allen C, Piror P, Hayward AC, editors. Bacterial wilt disease and the Ralstonia solanacearum complex. APS press, Minnesota USA; 2005. p. 9–28.
26. Tahat MM, Sijam K. Ralstonia solanacearum: The Bacterial Wilt Causal Agent. Asian J Plant Sci. 2010;9(7):385–93.
27. Fegan M, Prior P. How complex is the “Ralstonia solanacearum species complex”? In: C. Allen, P. Prior, and C. Hayward, editor. Bacterial Wilt: the Disease and the Ralstonia solanacearum Species Complex. Am Phytopathol Soc Press, St. Paul, MN.; 2005. p. 449–62.
28. Guidot A, Elbaz M, Carrère S, Siri MI, Pianzzola MJ, Prior P, et al. Specific genes from the potato brown rot strains of Ralstonia solanacearum and their potential use for strain detection. Phytopathology. 2009;99:1105–12.
29. Salanoubat M, Genin S, Artiguenave F, Gouzy J, Mangenot S, Arlat M, et al. Genome sequence of the plant pathogen Ralstonia solanacearum. Nature. 2002;415(6871):497–502.
30. Luo H, Lin Y, Gao F, Zhang C-T, Zhang R. DEG 10, an update of the database of essential genes that includes both protein-coding genes and noncoding genomic elements: Table 1. Nucl Acids Res. 2014;42(D1):D574–80.
31. Huang Y, Niu B, Gao Y, Fu L, Li W. CD-HIT Suite: a web server for clustering and comparing biological sequences. Bioinformatics. 2010;26(5):680–2.
32. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Bio. 1990 Oct;215(3):403–10.
33. Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007;35:W182–5.
34. Gupta A, Kapil R, Dhakan DB, Sharma VK. MP3: A Software Tool for the Prediction of Pathogenic Proteins in Genomic and Metagenomic Data. Raghava GPS, editor. PLoS One. 2014;9(4):e93907.
35. Yu NY, Wagner JR, Laird MR, Melli G, Rey S, Lo R, et al. PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics. 2010;26(13):1608–15.
36. Yu C-S, Cheng C-W, Su W-C, Chang K-C, Huang S-W, Hwang J-K, et al. CELLO2GO: A Web Server for Protein subCELlular LOcalization Prediction with Functional Gene Ontology Annotation. Raghava GPS, editor. PLoS ONE. 2014 Jun 9;9(6):e99368.
37. Chandra N. Computational approaches for drug target identification in pathogenic diseases. Exper Opin Drug Disc. 2011;6(10):975–9.
38. Hossain T, Kamruzzaman M, Choudhury TZ, Mahmood HN, Nabi AH, Hosen MdI. Application of the Subtractive Genomics and Molecular Docking Analysis for the Identification of Novel Putative Drug Targets against Salmonella enterica subsp. enterica serovar Poona. BioMed Res Int. 2017;2017:1–9.
39. Georrge JN, Umrania V. In silico identification of putative drug targets in Klebsiella pneumonia MGH78578. Ind J Biotechnol. 2011;10:432–9.
40. Sivashanmugam M, Nagarajan H, Vetrivel U, Ramasubban G, Therese KL, Narahari MH. In silico analysis and prioritization of drug targets in Fusarium solani. Med Hypotheses. 2015 Feb;84(2):81–4.
41. Allen C, Bent A, Charkowski A. Underexplored niches in research on plant pathogenic bacteria. Plant Physiol. 2009;150(4):1631–7.
42. Silver LL. Challenges of antibacterial discovery. Clin Microbiol Rev. 2011;24(1):71–109.
43. Galperin MY, Koonin EV. Searching for drug targets in microbial genomes. Curr Opin Biotechnol. 1999 Dec;10(6):571–8.
44. Folador EL, de Carvalho PVSD, Silva WM, Ferreira RS, Silva A, Gromiha M, et al. In silico identification of essential proteins in Corynebacterium pseudotuberculosis based on protein-protein interaction networks. BMC Syst Biol. 2016;10(1):103.
45. Hosen MdI, Tanmoy AM, Mahbuba D-A, Salma U, Nazim M, Islam MdT, et al. Application of a subtractive genomics approach for in silico identification and characterization of novel drug targets in Mycobacterium tuberculosis F11. Interdiscip Sci Comput Life Sci. 2014 Mar;6(1):48–56.
46. Maranhão FCA, Paião FG, Fachin AL, Martinez-Rossi NM. Membrane transporter proteins are involved in Trichophyton rubrum pathogenesis. J Med Microbiol. 2009;58(2):163–8.
47. Rodriguez GM, Smith I. Identification of an ABC Transporter Required for Iron Acquisition and Virulence in Mycobacterium tuberculosis. J Bacteriol. 2006;188(2):424–30.
48. Chen HD, Groisman EA. The Biology of the PmrA/PmrB Two-Component System: The Major Regulator of Lipopolysaccharide Modifications. Annu Rev Microbiol. 2013 Sep 8;67(1):83–112.
49. Butt AM, Nasrullah I, Tahir S, Tong Y. Comparative Genomics Analysis of Mycobacterium ulcerans for the Identification of Putative Essential Genes and Therapeutic Candidates. Carlow CK, editor. PLoS ONE. 2012;7(8):e43080.
50. Sperandeo P, Martorana AM, Polissi A. Lipopolysaccharide biogenesis and transport at the outer membrane of Gram-negative bacteria. Biochimica et Biophysica Acta (BBA) - Mol Cell Biol Lipids. 2017;1862(11):1451–60.
51. Li Y-H, Tian X. Quorum Sensing and Bacterial Social Interactions in Biofilms. Sensors. 2012;12(3):2519–38.
52. Sintim HO, Smith JA, Wang J, Nakayama S, Yan L. Paradigm shift in discovering next-generation anti-infective agents: targeting quorum sensing, c-di-GMP signaling and biofilm formation in bacteria with small molecules. Future Med Chem. 2010 Jun;2(6):1005–35.
53. Clatworthy AE, Pierson E, Hung DT. Targeting virulence: a new paradigm for antimicrobial therapy. Nat Chem Biol. 2007;3(9):541–8.
54. Band V, Weiss D. Mechanisms of Antimicrobial Peptide Resistance in Gram-Negative Bacteria. Antibiotics. 2014;4(1):18–41.
|






 This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License