IJCRR - 7(24), December, 2015
Pages: 43-47
Date of Publication: 20-Dec-2015
Print Article
Download XML Download PDF
CONSTRUCTION AND ANALYSIS OF MYOPATHY AND PARKINSON DISEASE PROTEIN NETWORK
Author: Sachin Rahangdale, Shobha Shouche, Ravikant Yadav, Harshad Sharma, Ankit Kumar Jain
Category: Healthcare
Abstract:Neurological Disorders are diseases of the central and peripheral nervous system. The disorders Parkinson's disease and myopathy are the traumatic disorders of the nervous system. The myopathies are neuromuscular disorders in which the primary symptom is muscle weakness due to dysfunction of muscle fiber and Parkinson's disease (PD) belongs to a group of conditions called motor system disorders, which are the result of the loss of dopamine-producing brain cells. Biological network is a network that applies to the biological system. These networks are widely used in many branches of biology as convenient representation of patterns of interaction between appropriate biological elements.Proteins-protein interaction (PPIs) in a cell form protein interaction networks (PINs) where proteins are nodes and their interaction are edges. In the present work we will investigate the network properties of proteins in disease protein-protein interaction network framework and identify common proteins between myopathy and Parkinson diseases.
Keywords: Framework, Interaction, Network, Edges, Neuromuscular
Full Text:
INTRODUCTION
Neurological Disorders are diseases of the central and peripheral nervous system.In other words, the brain, spinal cord, cranial nerves, peripheral nerves, nerve roots, autonomic nervous system, neuromuscular junction, and muscles are responsible for neurological disorders. These disorders include epilepsy, Alzheimer disease, myopathy and other dementias, cerebrovascular diseases including stroke, migraine and other headache disorders, multiple sclerosis, Parkinson’s disease, neuro-infections, brain tumours, traumatic disorders of the nervous system such as brain trauma, and neurological disorders as a result of malnutrition.[11] Hundreds of millions of people worldwide are affected by neurological disorders. Approximately 6.2 million people die because of stroke each year; over 80% of deaths take place in low- and middleincome countries. More than 50 million people have epilepsy worldwide. It is estimated that there are globally 35.6 million people with dementia with 7.7 million new cases every year - Alzheimer’s disease is the most common cause of dementia and may contribute to 60–70% of cases.[13] The Parkinson’s disease and myophaty on the traumatic disorders of the nervous system. The Myopathies are neuromuscular disorders in which the primary symptom is muscle weakness due to dysfunction of muscle fiber. Other symptoms of myopathy can include muscle cramps, stiffness, and spasm. Myopathies can be inherited (such as the muscular dystrophies) or acquired (such as common muscle cramps). Parkinson’s disease (PD) belongs to a group of conditions called motor system disorders, which are the result of the loss of dopamine-producing brain cells. The four primary symptoms of PD are tremor, or trembling in hands, arms, legs, jaw, and face; rigidity, or stiffness of the limbs and trunk; bradykinesia, or slowness of movement; and postural instability, or impaired balance and coordination. As these symptoms become more pronounced, patients may have difficulty in walking, talking, or completing other simple tasks. PD usually affects people over the age of 60. [12] Biological network is a network that applies to the biological system. These networks are widely used in many branches of biology as convenient representation of patterns of interaction between appropriate biological elements. [14]Proteinsprotein interaction (PPIs) in a cell form protein interaction networks (PINs) where proteins are nodes and their interaction are edges. The present study is designed to investigate the network properties of proteins in diseases, identification of common protein and their structural properties and protein-protein interaction network framework between myopathy and Parkinson diseases.
DISCUSSION
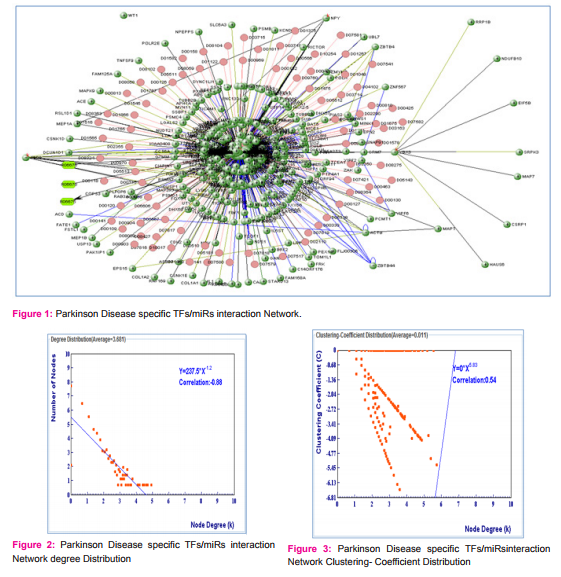
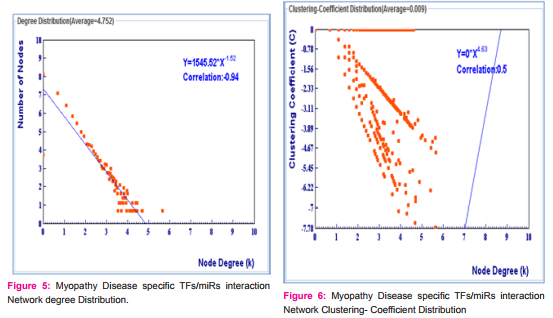
We have collected 5 miRNAs for Parkinson Disease and 31 miRNAs for Myopathy by using literature mining such as PubMed, miR2disease database. (Table-1) Using miRNA target database mirWalk, miRecords, miReg and miRTarBase. We have identified 298 targets for parkinson Disease and 1602 targets for myopathy disease. We made an interaction network using target proteins of miRs with the help of Visant. We have found the Degree of Distribution (Fig. 1, 2, 3) and Clustering- Coefficient Distribution of target proteins. (Fig 4, 5, 6) There are 131 proteins which are common between two diseases. It seems that these proteins are hotshots in these diseases, so required much more attention. We have plot a degree distribution of disease protein and the results indicate that most of the proteins are having 4 degrees. This is an interesting finding. It means that networks are obeying power law and they are scale-free in nature. The power laws are often interpreted as signatures of hierarchy and robustness.
ABCC1, ACTB, ACTG1, AKT1, ALDH1A1, APC, ARC, ARCN1, ARPP-21, ASAHL, ATP11C, BCL2, BCL2L11, BMP2, ,CBS, CBX5, CCND1, CDKN1B, CDKN1C, CEBPA, CNN3, CTGF, CXCL12, DGCR8, DICER1, DMPK, E2F1, E2F3, EGF, EGFR, EPHB2, ERBB2, ERG, ETS2, EXOSC1, FASLG, FASN, FBLIM1, FGFR1, FLT3, GALNT1, GAPDH, GRB2, HCN2, HDAC4, HELLS, HIST1, H2BB, HLA-G, HMGB1, HMOX1, HOXA9, HOXD10, HSPA1B, HSPB6, IARS, IDH1, IGF1, IGF1R, IGF2R, IL6, IMPACT, IRS1, ITGB1, ITLN1, JAG1, KLF4, KRT7, LIN28, LRRFIP1, LRRK2, MBL2, MCL1, MEF2A, MET, MLC1, MOCOS, MSI2, MYC, MYOD1, NFKB1, NFYC, NKX2- 3, NOTCH1, NPM1, NXN, PAK1, PAX3, PAX6, PAX7, PDE3A ,PIK3CA, PIWIL1, POLA2, POLR2A, POMC, PROC, PTBP1, PTBP2, PTEN,P TGS2, RAB34, RAC1, RDH10, RELB, RHOA, RHOD, RNASEN, RPE, RPL27, RUNX2, SLC11A1, SLC2A4, SLC7A1, SMAD2, SMAD5, SNCA, SOX2, SOX4, SRF, TFB1M, TGFB1, TGFBI, TGFBR2, TIAL1, TIMP2, TLR4, TLR9, TPPP, VEGFA, WT1
By doing functional annotation we found that all proteins which are common in these two diseases are reported in these following neurological pathways. (Table-2)
CONCLUSION
This study illustrates the analysis of Myopathy and Parkinson diseases. For this we have designed the protein-protein interaction analysis because each and every protein which involves in this study has their own role and importance and help to understand how Proteins are distributed in this network. We have identified about 131 common proteins which are hotshots for these two diseases and required much more attention to find out the drug target so that farther study can be designed to obtain the inhibitors of these targets.
ACKNOWLEDGMENT
Authors acknowledge the immense help received from the scholars whose articles are cited and included in references of this manuscript. The authors are also grateful to authors / editors / publishers of all those articles, journals and books from where the literature for this article has been reviewed and discussed. The authors would also like to thank Dr. Usha Shrivastav, Principal at Govt. M.V.M Ujjain and Mr. Krishna Kant Gupta for their support.
References:
1. Jiang Q., Wang Y., Hao Y., Juan L., Teng M., Zhang X., Li M., Wang G., Liu Y., (2009) miR2Disease: a manually curated database for microRNA deregulation in human disease. Nucleic Acids Res 37:D98-104.
2. Hsu SD, Lin FM, Wu WY, Liang C, Huang WC, Chan WL, Tsai WT, Chen GZ, Lee CJ, Chiu CM, Chien CH, Wu MC, Huang CY, Tsou AP, Huang HD. miRTarBase: a database curates experimentally validated microRNA-target interactions. Nucleic Acids Res. 2011 Jan;39(Database issue):D163-9. Epub 2010 Nov 10.
3. Dweep H, Sticht C, Pandey P, Gretz N. miRWalk--database: prediction of possible miRNA binding sites by “walking” the genes of three genomes.J Biomed Inform. 2011 Oct;44(5):839- 47. Epub 2011 May 14.
4. Xiao F, Zuo Z, Cai G, Kang S, Gao X, Li T. miRecords: an integrated resource for microRNA-target interactions. Nucleic Acids Res. 2009 Jan;37(Database issue):D105-10. Epub 2008 Nov 7
5. Barh D, Bhat D, Viero C. miReg: a resource for microRNA regulation. J Integr Bioinform. 2010 Aug 6;7(1). doi: 10.2390/ biecoll-jib-2010-144.
6. Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nature Protoc. 2009;4(1):44-57
7. Chen J, Bardes EE, Aronow BJ, Jegga AG. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009 Jul;37(Web Server issue):W305-11. Epub 2009 May 22.
8. Breitkreutz BJ, Stark C, Tyers M. Osprey: a network visualization system. Genome Biol. 2003;4(3):R22. Epub 2003 Feb 27.
9. Hu Z, Hung JH, Wang Y, Chang YC, Huang CL, Huyck M, DeLisi C. VisANT 3.5: multi-scale network visualization, analysis and inference based on the gene ontology. Nucleic Acids Res. 2009 Jul;37(Web Server issue):W115-21. Epub 2009 May 21.
10. Debmalya Barh1*, Neha Jain 1, 2, Triantafillos Liloglou3, Cedric vireo4, Kenneth Blum1,5, Vasco Azevedo6, Anil Kumar2, John K. Field3Identification of early biomarkers and molecular events in lung cancer using a novel reverse-transcriptomics approach.
11. National institute of Neurological Disorders and National Stroke, http://www.ninds.nih.gov/disorders/myopathy/myopathy.htm
12. Parkinson’s Disease Foundation,http://www.pdf.org/about_pd http://www.ucsfhealth.org/conditions/neurological_disorders
13. WHO,http://www.who.int/features/qa/55/en/
14. M. E. J. Newman, Networks: An Introduction, Oxford University Press, Oxford(2010). ISBN 0-19-920665-1


|






 This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License