IJCRR - 13(3), February, 2021
Pages: 43-49
Date of Publication: 03-Feb-2021
Print Article
Download XML Download PDF
Gene Expression Regulation by Epigenetic Mechanism an Emerging Way in Alcoholics
Author: Anjali Vagga, Ajay Meshram, Lata Kanyal, Komal Meshram
Category: Healthcare
Abstract:Environmental factors can impose most of their harmful effects, including harmful substances and drugs, by modifying the usual sequence of genes, advances to unusual expression or attempting to silence the major DNA sequences and their encoded proteins. Epigenetic is the research of modifications of DNA sequences and subsequent proteins, with no modification of the nucleotide sequences, Methylation of the DNA, alteration in Histone and RNA-mediated targeting control. Many biochemical reactions influences in human cell function, a pattern of their modification occurs in alcoholics. This review is focused on the regulation of gene expression by the epigenetic mechanism in alcoholics. Chronic consumption of alcohol in humans affects the brain, bringing the modifications in the expression of genes. Chronic alcohol intake also creates the desire for alcohol, loss of control, ultimately leading to liver damage. Such changes result in the expression of gene and cell cycle disturbance, facilitating the development of cirrhosis and cancer of the liver. DNA methylation, histone modifications and miRNA are the important common epigenetic modifications used as biomarkers of alcoholic liver disorders. While we are in the early stages of understanding the complex epigenetic regulatory system, the preliminary evidence provided here indicates that we could be at the dawn of the advancement of epigenetic factors in the diagnosis of various disorders.
Keywords: Epigenetic, Alcoholics, DNA methylation, DNA hypomethylation, Histone modification and MicroRNA
Full Text:
INTRODUCTION
Features of Alcoholic disease (ALD) includes inflammation, hepatic steatosis and deposition of fat that brings with it cirrhosis and hepatocellular carcinoma.1 Cognitive and affective states influence the pathogenesis of alcoholism which promotes the alcohol intake, might be due to different allostatic variables in multiple areas of the brain.2-4 Different researches have demonstrated the genetic and or environmental factors have played a key role in the development of alcoholism.5-7 It is proved that chronic consumption of Alcoholism is a replacing the brain disorders, chronic consumption of alcohol is characterized by loss of control while limiting alcohol intake, gets a negative emotional state while withdrawal and making the desire to seek alcohol.8,9 In animal models and humans, frequent alcohol consumption causes widespread brain changes in gene regulation.
Most of these contribute to cellular adaptations which at the end lead to dependence on alcohol and psychological tolerance. It is appreciated for the role of epigenetic changes in alcohol-induced modifications of expression and behaviour of genes for chronic alcoholism. e.g., chronic alcohol consumption results towards the shift in methylation of histone, DNA and Micro RNA transcription with different gene response to various brain cells types (e.g. glia and neurons) and proceeds to malfunction of cognitive function and plasticity of the brain involving the abuse of alcohol and dependence over it.10 Environmental conditions can impose most of their harmful effects, including harmful substances and drugs11, by modifying the usual epigenetic patterns, advancing to unusual expression or attempt to silencing the important sequences of DNA and their encoded proteins. Alcohol is quickly evolving as one of the key elements for changing the epigenome of cells and tissues from across organisms.12
What is epigenetics?
The term "epigenetic" simply means "in addition to changes in the genome." The concept has expanded to include all mechanism that involves the expression of a gene without modifying the sequence of DNA and advances to alterations which can be incorporated to new nucleoli (although studies indicate that certain genetic mutations can be altered). Exactly what the word will possibly continue to be discussed. Alterations in DNA which do not modify the sequence of DNA might alter the expression of genes. Chemical substances applied to separate genes may control their functioning; these changes are called epigenetic alterations. The epigenome contains all the chemical blends applied to the whole of particular DNA (genome) as a form of controlling its function (about expression) of entire genes in the genome. The epigenome's chemical blends are not a part of the particular DNA code, but are on or added to the DNA, epigenetic changes occur as cells divide, and may often be inherited through the gene.
Epigenetic modifications will boost to evaluate may be the genes are switched on or off, and which can impact protein synthesis in certain cells, assure that only proteins are synthesized that are required. For eg. The proteins are not synthesized in muscle cells, which help the growth of bone. Epigenetic change of pattern varies amongst individuals, growing tissues of the individual and also various cells.
Epigenetic modification
Histone modification
Post-translational modification of histone which involves the incorporation of phosphorus, incorporation of ubiquitin, methylation, Small Ubiquitin-like Modifier (SUMO) proteins are attached to or detached from other proteins and acetylene. The PTM formed for histones can vary gene expression by modifying the genetic makeup of chromatins or by including histone modifiers. Histone proteins work for inserting DNA into the chromosomes, DNA gets wrapped around eight histones. Histone alterations work in different biological procedures. Such as to the activation or deactivation the synthesis of mRNA, packing in the chromosome and genes repair or damage. Generally for most of the animal's incorporation of acetic acid to histone H3 is done with lysine 9, 14, 18, 23 and 56, methylation with Arginine 2 and Lysine 4,9,36 and 79 and phosphorylation at Ser 10 and 28, Thr 3 and 11. Acetylation of Histone H4 occurs with residues of Lysine at positions 16, 8, 5, 16, methylation of the Arginine at position 3, and of the lysine at position 20 and Phosphorylation of the serine at position 1.
To quantify particular modification of histone may provide good information for better understanding of the epigenetic regulation, cellular mechanism and the production of enzyme-based drugs to modify Histones.
Acetylation and Deacetylation of Histone
Incorporation of acetic acid to histone is done by the addition of acetyl groups from CoA. Incorporation of acetyl groups to Histone (Figure 1) has importance in maintaining different mechanisms inside the cell involving gene expression, repair of DNA, chromatin mechanics and transcription, nuclear import, repression of neurons, apoptosis, and progression of the cell cycle, DNA replication and differentiation. Enzymes Histone acetyltransferases (HATs) carry out the incorporation of the acetyl group to Histones and has an important role to regulate the incorporation of acetyl groups to Histones H3 and H4. About 20 HATs are known and are categorized as five families like GNAT 1, TAFII 250, P300/CBP MYST and co-activators like ACTR for nuclear receptors.13 Histone deacetylases (HDACs) inhibition increases Histone H3 Acetylation and inhibition of cap decreases the acetylation. Hydrolytic separation of acetyl groups from lysine residue of histone is done by enzymes named Histone Deacetylases (HDACs). Cancer development and genesis of Tumor is linked with an imbalance in Histone acetylation equilibrium. Histone H3 acetylation of the Lysine residue analysis can confer important knowledge to identify sites or patterns of acetylation, which provides deeper lights on epigenetic regulation of activation of the genes and production of HAT based drugs. Like that of HATs, HDACs also have key roles in particular mechanisms involving Histone H3 and H4. So far four groups of HDACs have been established. 1,2,3 and 8 are included in class I HDAC, Class II HDAC include 4, 5, 6, 7, 9 and 10, Class III includes molecules like sirtuins which requires a cofactor with NAD+ and comprises of SIRTs 1-7 and a class IV group of enzymes includes HDAC 11 and has both classes I & II functionality. By inhibiting HDAC gets impact on the cellular mechanisms, apoptosis and differentiation of the cancer cells. HDAC inhibitors act like anticancer agents.14
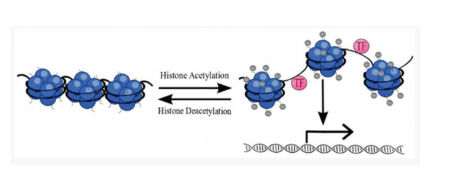
Figure 1: Represent the reactions of acetylation and deacetylation. In the acetylation process relaxation of chromatin structure with which TF bind easily. Deacetylation leads to chromatin inactivation.15
Histone Methylation and Demethylation
Addition of methyl group to Histone is a phenomenon carried out by histone methyltransferases (HMTs) comprising either 1, 2 or 3 CH3 units derived from SAM to Lys or Arg residues of Histones. Methylation of DNA is regulated by HMTs by the processes activation or repression of transcription in the chromatins. Methylation in the Histone occurring in the cell nucleus, different genes of the DNA, which are complexed with histone can be suppressed or activated16. Arginine and Lysine residues which are modified by histone methyl transferases are of different types e.g. SET 1, SET 7/9, Histone H3, ALL-1, Ash 1, ALR, MLL, SMYD3 and Trx are Histone methyltransferases which brings the transfer of methyl group to Histone H3 in mammalian cells at Lys 4 (H3-K4).
Enzyme Histone methyltransferases like SUV 39- h1, SUV 39- h2, Dim -5, G9a and Eu- HMT brings the transfer of CH3 group to Histone H3 in mammalian cells at Lys 9 (H3-K9). Histone methyltransferase enzymes like EZH2 and G9a brings the CH3 group transfer to Histone H3 in mammalian cells at Lys 27 (H3-K27)17. CH3 group transfer to H3-(K9 and K27) helps in the synthesis for heterochromatin and also plays a role in signals for silencing of gene expression over the euchromatic sites. Increase in H3-K27 methylation amount globally is also found to be associated with a pathological condition like the progression of cancer.
DNA Methylation
The commonest form of epigenetic alteration is one, i.e. DNA methylation. There is the attachment of small molecules in DNA methylation, comprising of one carbon atom three hydrogen atoms, to DNA segments, called methyl groups. When a specific gene is attached by methyl group that gene is turned off or can say it is silenced, then no protein has formed that gene. DNA methylation is the so far most studied epigenetic mechanism which occurs by cytosine modification by covalent means of joining CH3 group to base at 5’ cytosine ring carbon located inside CpG dinucleotides. CpG dinucleotides more than 85% in number, are spread across the genome and are present in respective sequences are highly hypermethylated/ transcription silenced in the normal cells, the condition which is critical to the integrity of the structure of genome chromatin.
A vital role is played by DNA methylation for recombining, repairing and replication of DNA and also in controlling the activity of genes. The addition of the H3 group to the sequence of DNA at the 5’ cytosine base is necessary for the formation of CpG dinucleotide. A DNA methyltransferase (DNMTs) family brings out this process. CpG rich areas form the area generally called as the island with pieces of 200 bp and can form kilobases in length, and situated near the promoter regions of strong expressing genes, and these are the prone areas for methylation in human tumours, like prostate tumours. Islands of CpG produce the complex subunits and the processes occurring like methylation and demethylation ultimately produces activation or inactivation in nearly 55% of the cases. That is why the methylations of the CpG islands in gene promoter region can prevent or de-regulate gene product synthesis.18-20
Three major forms of DNMTs like (DNMT1, DNMT3A, and DNMT3B) are identified. DNMT2 known as the fourth enzyme has previously been not a methyltransferase of DNA. A clear sequence is present in DNMT3 or TRDMT 1 similarly with 5- methylcytosine methyltransferases, but methyl group position 38 in aspartic acid transfer RNA has been shown as enzyme and doesn’t methylated DNA. There is the belief that a small quantity of mammalian DNMT2 is referred to in recombining DNA, recognizing DNA damage and repairing mutations. DNA methylation patterns are preserved by responsible enzyme DNMT1.
At the replication fork, DNMT 1 is situated. Newly formed DNA gets methylated to DNMT3 A/B and cannot distinguish among unmethylated and hemimethylated CpG sites and they can’t copy a common trend of the CH3 group transfer or help to sustain the trend of CH3 transfer.21 When the DNA gets methylated, condensation of chromatin occurs and then the complex of transcriptions is not able to bind DNA and thereby silences gene expression. This type of proteins, in effect, involves the enzymes capable for further epigenetic changes which lead to condensed chromatin state.22,23
DNA hypomethylation
Most studies have indicated different triggers for DNA hypomethylation, including lack of precursors of S-adenosylmethionine or vitamin-like folic acid in the food or defect in the gene of the metabolic pathway for the CH3 group donor. A deficit of enzyme methyltransferase can cause hypomethylation of DNA. Hypomethylation of the sites called promoter regions of the genome may result in the decrease in the stability of the genome also by increasing the expression of transposons, which remain dormant due to methylation under normal physiological conditions. Low levels of methylations may result in lower stability of chromosome and lower activation of proto-oncogene.18-20
Eukaryotic genomes structural and functional organization is reflected by Chromatin24,25 and it is composed of RNA, DNA and variety of protein components.24 Nucleosome comprising146 bp is the main repeat unit of chromatin; it is placed such that it wraps around core histones H2A, H2B, H3 and H4 and then forms an octamer.26 Nuclear protein Histone acyl transferases (HATs) can reverse Histone tails which contain residues of amino acids, Histone lysine methyltransferases (KMTs), Histone deacetylases (HDACs) and Kinases are examples.27-29
Mechanism of epigenetic
During development, the prediction is that pattern of CpG methylation changes. During the development of the embryo, methylation is wiped out throughout the genome and then it is restored to all but CpG bunches (genome sites with dense CpG residues). Until some of the CpG bunches are methylated otherwise they remain hypomethylated later during the development. 30,31 Transcriptional repression is associated with further transfer of CH3 group to cyt in CpG groups and other similar CpG dinucleotides,31-33 particularly when such methylated sites include promoters or some different sites of gene regulation.33 Nonetheless, transfer of methyl group to DNA could be activated if it inhibits transcriptional repressors from being bound or restricts their expression. Recent research in mammalian promoters that define the degree of methylation indicates that methylation takes place at over a small quantity in the proportion of CpG dinucleotides and transcription inhibition occurs at only small regions of genes in differentiated cells, some of the repressed lengths of genes are particular to germline2, like the pluripotency genes, indicating that methylation is a critical mechanism for the suppression of main genes during their separation.32
Epigenetic Modification in Alcoholics
Stress and alcoholism can contribute to changes in epigenetic mechanism and that can be associated with behavioural phenotypes and synaptic remodellings like depression and anxiety.34 At the outset, we will outline the molecule that regulates the synaptic plasticity, which is known to associate with the addiction of alcohol and disorders of stress and then we will overview their regulation by an epigenetic mechanism, specifically the DNA methylation and Histone acetylation/methylation, which underlies these disorders. However, epidemiological findings of the effects of associated drinking and smoking, there is an indication that epigenetic processes are doubtful about alcohol consumption rather than to smoking. Heavy drinking (>80 g/d) and smoking one pack per day together raises the risk of cancer of oesophagus by up to 44 times.35 An epidemiological finding clearly indicates there is a distinct toxicity process of drinking and smoking which will confer the disease risk.
Likewise, it is shown that the risk of cancer and other smoking-related disorders are not only due to cigarette smoking, but other the associated effects of various toxic materials found in smoke, alcohol-related risks are linked to dosage and alcohol content, with the concentration of alcohol on the higher side, confers increased risk.36 From the viewpoint of the epigenetic mechanism, looking at observations, researchers carried out the specific evaluation of both candidate genes and deeper investigations using advanced array-based analysis platforms for the evaluation of fundamental processes at hand. The utility of current alcohol biomarkers is limited to.37 Measuring alcohol in the breath or serum will be the best bio-indicator for alcohol. Present an analysis determines the actual consumption of alcohol at present and do not differentiate acute consumption and chronic violence. Alcoholism has some biomarkers of adverse effects like specificity and sensitivity, which are not used as screening tools. Biomarkers are required, looking at the magnitude of health problems and community costs associated with drinking alcohol.
DNA methylation in alcoholic
For DNA methylation S adenosyl methionine acts as a predominant methyl donor, in alcoholics, it is deficient, which leads to hyperhomocysteinemia which also occurs commonly in ALD patients.38 DNA methylation in alcoholics is also affected by reducing the amount of SAM. In the experiments over Rats, it is found that IV ingestion of alcohol diet for 9 weeks showed a drastic reduction in glutathione, SAM, Methionine and reduced methylation of DNA.39 DNA hypomethylation can advance to structural change in chromatin and expression of genes which may lead to strand breaks and DNA damage.39,40 which forms the environment for malignancy.41 In hepatocellular carcinoma, there is a strong correlation between alcohol consumption and reduced methylation of gene O6- methyl guanine DNA methyltransferase. It is shown by researchers that it is the alcohol-metabolizing enzyme, ADH1.
Alcoholic modifications of Histone
Phosphorylation, acetylation and transfer of CH3
In vivo and in vitro there is research evidence that alcohol consumption brings the epigenetic changes in organs like gastrointestinal system, liver and brain.42 Alcohol consumption brings out the effect of liver Histone acetylation/ methylation and phosphorylation. Specifically, transfer of acetyl groups to Histone H3 at Lys 9 (H3AcK9) has been reported in exposed Iry rat hepatocytes which were subjected to alcohol in vitro.43 Other lys residues like H3 lys 14, lys 18 and lys 23 were not acetylated. It is observed that alcohol consumption modulates H3 acetylation by increasing HAT activity and inhibition of HDAC.44 Acetylation of Histone depends on the HAT and HDAC activities.45 In some cases, the HAT / HDAC balance controls the Histone residues which are getting acetylated and regulation of gene expression.46 Consumption of alcohol seems to alter the function of HAT and HDAC in hepatocytes.43 Hepatic cell exposure to in vitro alcohol affects the role of HDAC6 which directly affects the dynamics of the microtubules.47 Liver cells exposed to alcohol show lower levels of Class III HDAC and sirtuin 1 (SIRT1) mRNA expression.48
Acetylation of Histone and methylation of DNA are involved in the transcription and/or silencing cycle of the genes in disease states.49 Generally, in target gene promoters, CpG islands having higher methylation leads to deacetylation of local histones, on the other hand, small quantities of transfer of acetyl groups to Histone appear to incite DNA for attachment of CH3 group. There is close co-operation of these two epigenetic pathways but is there any hierarchical order of the incidence is still not clear. In chronic alcoholic disorders, there is no connection between hyperacetylation of H3K9, elevated methylation of H3K4 and loss of methylation along with global DNA hypomethylation. Research carried out for interaction between epigenetic events can provide the required information for ALD mechanism.
MicroRNA
The small non-coding RNAs Micro RNAs (miRNAs) controls various physiologic and pathologic activities at the level of post- transcription by modulating gene expression. Different pieces of evidence provided the importance of miRNA in further advancement and severity of the diseases of the liver. Most of the studies analyzed the effect of miRNA on Alcoholic liver disease (ALD) and Non-alcoholic Fatty liver disease (NAFLD)50, which shares a similar underlying mechanism and pathological characteristics.
In reality, all the pathological and physiological processes involve human miRNAs, including signal transduction, cell proliferation and differentiation, viral host interaction, metabolism, oncogenesis and inflammation and immune response.51-52 The expressions of a wide range of miRNAs are controlled by several factors, like cigarette smoking, alcohol and diet and some drugs.53 Now miRNA has gained importance as one of the main factors for identifying the cause of various diseases and as potential biomarker therapeutic target and for diagnosis.52 Considering the fast investigations and analysis of the role of miRNAs over the last few years, an updated overview of the subject will be looked after first, followed by a note on miRNA changes common to both Non-Alcoholic Fatty Liver Disease and Alcoholic Liver Disease.
Role of micro-RNA in alcoholics
The occurrence of the various types of ALD like steatosis, alcoholic hepatitis and cirrhosis comprise chronic alcoholics and heavy drinkers along with the probability of disease. The underlying pathophysiology for ALD is based on the direct cytotoxic impact of alcohol intake and the change in inflammatory response influenced by ethanol.54 Enzymes like cytochrome P4502E1 (CYP2E1) and alcohol dehydrogenase (ADH) contributes for alcohol metabolism55 which leads to oxygen free radicals, acetaldehyde and nitric oxide, which in the flow may cause cellular damage and inflammation of the liver.56 Toxicity of acetaldehyde causes an increase in bacterial lipopolysaccharide (LPS) permeability to the intestine, which joins to receptors-4 (TLR-4) and incites stellate cells and Kupfer cells via pro-inflammatory cytokines, like tumour necrosis factor α - (TNF).57 The transmission of the inflammatory signal is through the pathway of the nuclear factor-KB (NF-KB), which proceeds to liver damage.58
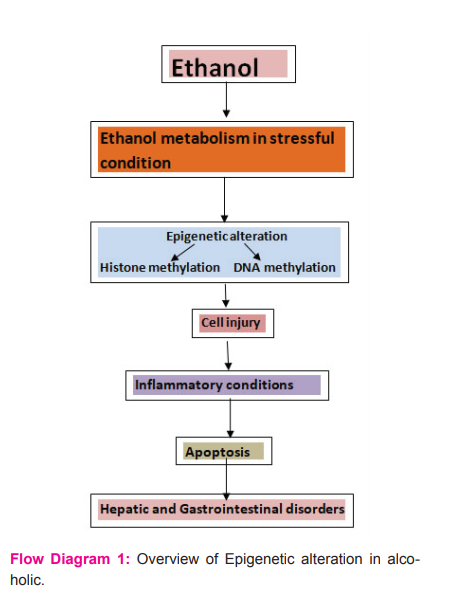
Conclusion
Epigenetics is the research of DNA alteration and accompanying proteins, without any change in the sequence of the gene. Many biochemical reactions are regulated by DNA methylation, histone alteration, and RNA-mediated targeting and they have the main role in cellular functioning, these are modified in alcoholic subjects. Alcoholism brings the changes in the expression of gene and cell cycle disturbance, facilitating the development of liver cancer like cirrhosis of the liver. DNA methylation, histone modifications and miRNA are the important common epigenetic modifications used as biomarkers of alcoholic liver disorders. While we are in the early stages of understanding the complex epigenetic regulatory system, the preliminary evidence provided here indicates that we could be at the dawn of the advancement of epigenetic factors in the diagnosis of various disorders.
Acknowledgment:
Nil.
Conflict of interest:
Nil
Source of funding:
Nil
References:
-
Mandrekar P. Epigenetic regulation in alcoholic liver disease. World J Gastroenterol 2011;17(20):2456.
-
Pandey SC. The gene transcription factor cyclic AMP-responsive element-binding protein: role in positive and negative affective states of alcohol addiction. Pharmac Ther 2004;104(1):47-58.
-
Koob GF. Alcoholism: allostasis and beyond. Alcoholism: Clin Expt Res 2003;27(2):232-243.
-
Koob G, Kreek MJ. Stress, dysregulation of drug reward pathways, and the transition to drug dependence. AmJ Psych 2007;164(8):1149-1159.
-
Edenberg HJ, Foroud T. The genetics of alcoholism: identifying specific genes through family studies. Addic Bio 2006;11(3?4):386-396.
-
Ducci F, Goldman D. Genetic approaches to addiction: genes and alcohol. Addiction 2008;103(9):1414-1428.
-
Farris SP, Wolen AR, Miles MF. Using expression genetics to study the neurobiology of ethanol and alcoholism. Int Rev Neurobiol 2010;91:95-128.
-
Koob GF, Volkow ND. Neurocircuitry of addiction. Neuropsychopharmacology 2010;35(1):217-238.
-
Koob GF. Neurocircuitry of alcohol addiction: synthesis from animal models. In Handbook of Clin Neurol 2014;125:33-54.
-
Ponomarev I. Epigenetic control of gene expression in the alcoholic brain. Alcohol Res Curr Rev 2013;35(1):69.
-
Ambili R. Drug resistance in cancer chemotherapy-An overview. Int J Curr Res Rev 2013;5(08):41-46.
-
Shukla SD, Zakhari S. Epigenetics- a new frontier for alcohol research. Alcohol Res: Curr Rev 2013:35(1):1.
-
Kuo MH, Allis CD. Roles of histone acetyltransferases and deacetylases in gene regulation. Bioessays 1998;20(8):615-626.
-
Dokmanovic M, Clarke C, Marks PA. Histone deacetylase inhibitors: overview and perspectives. Mol Cancer Res 2007;5(10):981-989.
-
Koprinarova, M.; Schnekenburger, M.; Diederich, M. Role of Histone Acetylation in Cell Cycle Regulation. Curr Top Med Chem 2015;16:732–744.
-
Greer EL, Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nature Rev Genet 2012;13(5):343-357.
-
Wood A, Shilatifard A. Posttranslational modifications of histones by methylation. In Advances in protein chemistry 2004; 67: 201-222). Academic Press.
-
Chin SP, Dickinson JL, Holloway AF. Epigenetic regulation of prostate cancer. Clin Epigene 2011;2(2):151-169.
-
Day TK, Bianco-Miotto T. Common gene pathways and families altered by DNA methylation in breast and prostate cancers. Endocrine-related Cancer 2013;20(5):R215-232.
-
Majumdar S, Buckles E, Estrada J, Koochekpour S. Aberrant DNA methylation and prostate cancer. Curr Genom 2011;12(7):486-505.
-
Subramaniam D, Thombre R, Dhar A, Anant S. DNA methyltransferases: a novel target for prevention and therapy. Front Oncol 2014;4:80.
-
Boyes J, Bird A. DNA methylation inhibits transcription indirectly via a methyl-CpG binding protein. Cell 1991;64(6):1123-1134.
-
Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, et al. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature 1998;393(6683):386-389.
-
Horn PJ, Peterson CL. Chromatin higher order folding--wrapping up transcription. Science 2002; 297(5588):1824-1827.
-
Yap KL, Zhou MM. Structure and function of protein modules in chromatin biology. InChromatin Dynamics in Cellular Function 2006 Jan 1 (pp. 1-23). Springer, Berlin, Heidelberg.
-
Rando OJ, Ahmad K. Rules and regulation in the primary structure of chromatin. Curr Opin Cell Bio 2007;19(3):250-256.
-
Mottet D, Castronovo V. Histone deacetylases: target enzymes for cancer therapy. Clin Expt Metast 2008;25(2):183-189.
-
Perez-Cadahia B, Drobic B, Khan P, Shivashankar CC, Davie JR. Current understanding and importance of histone phosphorylation in regulating chromatin biology. Curr Opin Drug Disc Dev 2010;13(5):613.
-
Shukla A, Chaurasia P, Bhaumik SR. Histone methylation and ubiquitination with their cross-talk and roles in gene expression and stability. Cell Mol life Sci 2009;66(8):1419-1433.
-
Shen L, Kondo Y, Guo Y, Zhang J, Zhang L, Ahmed S, et al. Genome-wide profiling of DNA methylation reveals a class of normally methylated CpG island promoters. PLoS Genet 2007;3(10):e181.
-
Illingworth RS, Bird AP. CpG islands–‘a rough guide’. FEBS letters 2009; 583(11):1713-1720.
-
Weber M, Hellmann I, Stadler MB, Ramos L, Pääbo S, Rebhan M, et al. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet 2007;39(4):457-466.
-
Bird AP. CpG-rich islands and the function of DNA methylation. Nature 1986;321(6067):209-213.
-
Moonat S, Pandey SC. Stress, epigenetics, and alcoholism. Alcohol Res Curr Rev 2012; 34(4):495–505.
-
Tuyns AJ, Esteve J, Raymond L, Berrino F, Benhamou E, Blanchet F, et al. Cancer of the larynx/hypopharynx, tobacco and alcohol: IARC international case?control study in Turin and Varese (Italy), Zaragoza and Navarra (Spain), Geneva (Switzerland) and Calvados (France). Int J Can 1988;41(4):483-491.
-
Pöschl G, Seitz HK. Alcohol and cancer. Alcohol Alcohol 2004;39(3):155-165.
-
Tavakoli HR, Hull M, Okasinski LM. Review of current clinical biomarkers for the detection of alcohol dependence. Innov Clin Neurosci 2011;8(3):26.
-
Lu SC, Martínez?Chantar ML, Mato JM. Methionine adenosyltransferase and S?adenosylmethionine in alcoholic liver disease. J Gastroenterol Hepatol 2006;21:S61-64.
-
Lu SC, Huang ZZ, Yang H, Mato JM, Avila MA, Tsukamoto H. Changes in methionine adenosyltransferase and S-adenosylmethionine homeostasis in alcoholic rat liver. Am J Physiol Gastroint Liver Physiol 2000;279(1):G178-185.
-
Pogribny IP, Basnakian AG, Miller BJ, Lopatina NG, Poirier LA, James SJ. Breaks in genomic DNA and within the p53 gene are associated with hypomethylation in livers of folate/methyl-deficient rats. Cancer Res 1995 May 1;55(9):1894-1901.
-
Martínez?Chantar ML, Corrales FJ, Martínez?Cruz LA, García?Trevijano ER, Huang ZZ, Chen L, et al. Spontaneous oxidative stress and liver tumors in mice lacking methionine adenosyltransferase 1A. FASEB J 2002;16(10):1292-124.
-
Shukla SD, Velazquez J, French SW, Lu SC, Ticku MK, Zakhari S. Emerging role of epigenetics in the actions of alcohol. Alcoholism: Clin Expt Res 2008;32(9):1525-1534.
-
Park PH, Miller R, Shukla SD. Acetylation of histone H3 at lysine 9 by ethanol in rat hepatocytes. Biochem Biophy Res Comm 2003;306(2):501-504.
-
Park PH, Lim RW, Shukla SD. Involvement of histone acetyltransferase (HAT) in ethanol-induced acetylation of histone H3 in hepatocytes: a potential mechanism for gene expression. Am J Physiol Gastroint Liver Physiol 2005;289(6): G1124-1136.
-
Ito K, Adcock IM. Histone acetylation and histone deacetylation. Mol Biot 2002;20(1):99-106.
-
Huber LC, Brock M, Hemmatazad H, Giger OT, Moritz F, Trenkmann M, et al. Histone deacetylase/acetylase activity in total synovial tissue derived from rheumatoid arthritis and osteoarthritis patients. Arthr Rheum 2007;56(4):1087-1093.
-
Shepard BD, Joseph RA, Kannarkat GT, Rutledge TM, Tuma DJ, Tuma PL. Alcohol?induced alterations in hepatic microtubule dynamics can be explained by impaired histone deacetylase 6 function. Hepatology 2008;48(5):1671-1679.
-
Lieber CS, Leo MA, Wang X, DeCarli LM. Effect of chronic alcohol consumption on Hepatic SIRT1 and PGC-1α in rats. Biochem Biophys Res Comm 2008;370(1):44-48.
-
Vaissière T, Sawan C, Herceg Z. Epigenetic interplay between histone modifications and DNA methylation in gene silencing. Mutat Res 2008;659(1-2):40-48.
-
Torres JL, Novo-Veleiro I, Manzanedo L, Alvela-Suárez L, Macías R, Laso FJ, et al. Role of microRNAs in alcohol-induced liver disorders and non-alcoholic fatty liver disease. World J Gastroenterol 2018;24(36):4104.
-
Ambros V. The functions of animal microRNAs. Nature 2004;431(7006):350-355.
-
Hammond SM. An overview of microRNAs. Adv Drug Del Rev 2015;87:3-14.
-
Dolganiuc A, Petrasek J, Kodys K, Catalano D, Mandrekar P, Velayudham A, et al. MicroRNA expression profile in Lieber?DeCarli diet?induced alcoholic and methionine choline-deficient diet?induced nonalcoholic steatohepatitis models in mice. Alcoholism: Clin Expt Res 2009;33(10):1704-1710.
-
Laso FJ, Pastor I, Orfao A. Immune system and alcoholic liver disease. Med Clin 2005;125(7):263.
-
Lieber CS. Hepatic and metabolic effects of ethanol: pathogenesis and prevention. Ann Med 1994;26(5):325-330.
-
Mello T, Ceni E, Surrenti C, Galli A. Alcohol induced hepatic fibrosis: role of acetaldehyde. Mol Aspects Med 2008;29(1-2):17-21.
-
Gao B, Bataller R. Alcoholic liver disease: pathogenesis and new therapeutic targets. Gastroenterology 2011;141(5):1572-1585.
-
Mandrekar P, Szabo G. Signalling pathways in alcohol-induced liver inflammation. J Hepatol 2009;50(6):1258-1266.
|






 This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License