IJCRR - 13(1), January, 2021
Pages: 146-149
Date of Publication: 05-Jan-2021
Print Article
Download XML Download PDF
Classification of Diabetes Using Deep Learning and SVM Techniques
Author: K. Thaiyalnayaki
Category: Healthcare
Abstract:Background: Diabetes is a disease increasingly alarming affecting people worldwide. If not treated properly, it affects organs. An automatic classification of diabetes using deep learning perceptron and SVM is attempted.
Objective: To identified problem and provide automatic classification of diabetics data and the solution using deep learning perceptron and SVM for the best therapeutic management.
Method: The dataset consists of 768 instances, out of which, 500 diabetes subjects and 268 healthy people.
Results: There are 9 attributes which are used for analysis. MLP deep learning classifier with 18 parameters are utilized to correctly classify 595 Instances with a classification accuracy of 77.474 % and Incorrectly Classified are 173 Instances with 22.526 %. The comparison of MLP deep learning classifier with SVM classifier is performed where the SVM classifier Correctly Classified Instances are 500 with a classification accuracy of 65.1042 % and Incorrectly Classified Instances are 268 with 34.8958 %.
Conclusion: Deep learning perceptron classifier performs well with diabetes dataset and can be used for further automatic identification and detection analysis.
Keywords: Diabetes, Deep learning, SVM, ROC, confusion matrix, RBF
Full Text:
INTRODUCTION
Diabetes is an illness using which, blood sugar is not metabolically processed in the physique. Its frequency rates are expanding alarmingly consistently. If untreated, diabetes-related complexities in numerous imperative organs of the body may turn destructive. Diabetes is the main reason for visual impairment and visual debilitation in grown-ups in developed nations and more than one million lower appendage removals every year.1 One of the promising methods in AI is Support Vector Machine (SVM). SVM is utilized for classification of system. PCA is utilized for reducing the measurements by keeping up a lot of difference however much as could reasonably be expected. Output yield from preprocessed information is presently accomplished as the input.2 Unsupervised and supervised learning methods are utilized for sampling and structure, which are then trailed by a rule-based algorithm. Genuine diabetes dataset show that coherent SVMs give a promising tool to the forecast of diabetes, where ruleset have been produced, with a good prediction accuracy.3 The accuracy accomplished by useful classifiers such as Artificial Neural Network, Naive Bayes method, Decision Tree and Deep Learning lies within the scope of 90–98%. Then it is meaningful to build up a framework in the form of an application or a site that can utilize the method to help social healthcare authorities in the early discovery of diabetes.4 All trials are run on GPU empowered TensorFlow with Keras structure. Highlights of profound learning system are explored, based on CNN-LSTM architecture and into SVM for classification. LSTM can deal with long haul conditions in a time series data. To choose two paths of investigation, the data were run for SVM with direct and RBF part.5
MATERIALS AND METHODS
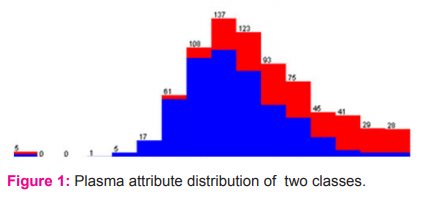
The dataset of 768 samples are from Pima Diabetes, National Institute of Diabetes and Digestive and Kidney Diseases. There are 9 attributes Plasma glucose concentration, 2 hours in an oral glucose tolerance test, Two-hour serum insulin in mu U/ml, Diastolic blood pressure in mm Hg, Body mass index, Diabetes pedigree function, Age, triceps skinfold thickness in mm, class variable as 0 or 1. Class desirability and the number of instances are 0 for 500 instances and 1 for 268 instances. Figure 1 represents the distribution of the second attribute. A few imperatives were put on the determination of these cases from a bigger database. Specifically, all-female patients are at any 21 years of age of Pima Indian legacy. ADAP is a versatile learning schedule that produces and executes advanced analogues of perceptron like gadgets.
RESULT AND DISCUSSION
The attribute data set is trained using Multilayer perceptron deep learning architecture. XAVIER Neural network configuration is used to train it to learn. Trainable parameters are 18.The output layer consists of 8 inputs, 2 outputs with W:{8,2}, b:{1,2}. Bias Initialization is first set 0, dropout of neurons is disabled. Adam is a versatile learning rate streamlining method that has been structured explicitly for training profound neural systems. It helps to find individual learning rates for each parameter. Adams updater is chosen with learning rate as 0.001, learning rate constant schedule value as 0.001 and beta1 as 0.9, beta2 as 0.999, epsilon as 1.0e-8. Bias updater for learning is also scheduled. Batch processing is performed with seed as 0 and stochastic gradient descent optimization Algorithm is set. Weight noise is not provided, hence disabled. The loss function is minimized and gradient normalization is performed with, gradient normalization threshold as 1. 10 fold cross-validation is applied. The training process includes
-
Parameters Initialization
-
optimization algorithm
-
Input is propagated to the network
-
Cost function computation
-
Gradient’s of the cost concerning parameters is calculated.
-
Each parameter is updated using the gradients concerning optimization algorithm.
The statistical analysis of classified instances are Kappa statistic is 0.4832, Mean absolute error is 0.317, Root mean squared error is 0.3951, Relative absolute error is 69.7509% and root relative squared error is 82.8893%. The Correctly Classified Instances are 595 with a classification accuracy of 77.474 % and Incorrectly Classified Instances 173 are 22.526% (Table 1 and 2).
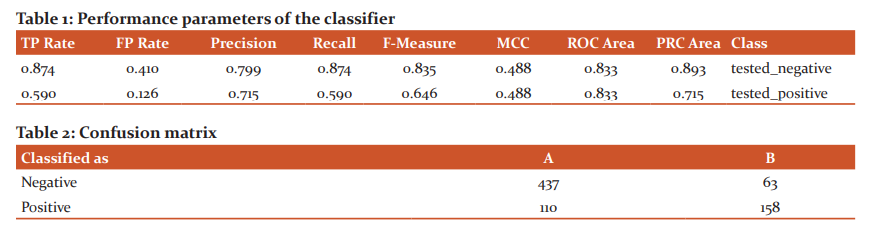
The lib SVM classifier is compared with the performance of deep learning architecture performance and classifier accuracy obtained for the pima dataset is 75% and the time is taken to build model is 0.13 seconds. Table 1 and 2 represents the Performance parameters and Confusion matrix of the classifier.
RBF Network
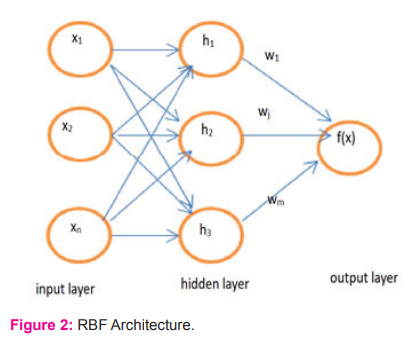
Radial Basis Function Network which constructs the hidden layer in an unsupervised procedure and RBF classifier which is entirely supervised. RBF Network makes a reality of a systematized Gaussian radial basis function network. It utilizes the k means clustering calculation to give the basis function and a linear regression is learned. Each cluster’s symmetric multivariate Gaussians are suitable for the data. On the off chance that the class is ordinary, it utilizes the given number of groups per class RBF regressor implements Gaussian radial basis function networks frameworks for regression, arranged in a coordinated way utilizing optimization class by restricting squared error with the BFGS strategy. In numerical optimization, the Broyden Fletcher Goldfarb Shanno (BFGS) system is an iterative computational procedure for taking care of unconstrained nonlinear streamlining issues.RBF architecture is shown in figure2. All traits are standardized into the [0,1] scale. It is possible to use conjugate gradient descent instead of BFGS refreshes, which is faster for cases with various boundaries, and normalized basis works as opposed to unnormalised ones.6-8
The inceptive pivot of the Gaussian radial basis functions discovered utilizing basic K Means. The ? assessed values are assigned to the maximum distance between any centre and its nearest neighbour in the set of centres.
There are various parameters in the network. The edge boundary is utilized to punish output layer weights. The number of premise functions can likewise be determined. Huge numbers produce long training times. Another alternative decides if one global sigma esteem is utilized for all units, quickest, regardless of whether one worth is utilized per unit and set as the default, or an alternate value is found out for each node mix. It is also possible to learn attribute weights for the distance function.9,10
The output shown is the squared value of the input. At long last, conjugate gradient descent can be utilized rather than BFGS updates, that is quicker for instance with numerous boundaries, and to apply standardized basis functions rather than unnormalized ones. An estimated form of the strategic function is utilized as the initiation work in the yield output layer to improve speed. Additionally, if delta esteems in the backpropagation step are inside the client described tolerance, the gradient isn't refreshed for that specific case, which spares several extra time. Parallel computation of squared error and gradient is conceivable when multiple CPU cores are available. Information is part of groups and prepared in different threads for this situation. Runtime for larger datasets is improved.
Nominal attributes are processed using the unsupervised model. Nominal To Binary filter and missing qualities are supplanted globally using replace missing values with choices. Batch size 100 is used. The seed is the random number seed to be used and set as 1.Use Normalized Basis Functions decides whether to use normalized basis functions or not. Scale optimization option gives the number of sigma parameters to use. The quantity of threads to utilize chooses a size of string pool. The attribute weights are used. decimal places concerning three positions are utilized for the output yield of model numbers. The batch size chooses the favoured number of cases to process if group prediction is being carried out. Approximately cases might be given, nevertheless allows executions to determine a favoured cluster size. The resilient parameter for the delta esteems is set as 1.0e-6. The ridge penalty factor for the output layer is decided as 0.01.poolSize is set as 1 which is the thread pool size, for instance, the CPU cores. The conjugate gradient descent is utilized which is suggested for some parameters. The number of premise functions to utilize is additionally set beforehand.8-10
The abilities are binary class, missing class esteems, nominal class, attributes are binary traits, date features, empty nominal qualities, missing values, nominal characteristics, numeric quality, unary characteristics, interfaces are randomizable, Handlers are weighted instances. Additional Minimum number of instance is set 1. There are several parameters. All parameters are set as described above. To enhance speed, a tentative form of the strategic function is utilized as the activation function in the yield layer. Likewise, if delta esteems in the backpropagation steps are inside the client indicated resilience, the gradient is not refreshed for that specific case, which spares some extra time. Equal estimation of squared error and slope is conceivable when numerous CPU centres are available. Information is part of clumps and handled in independent strings for this situation. Note this just improves runtime for bigger datasets. Nominal traits are handled utilizing the unsupervised nominal to binary channel and missing qualities are supplanted all-inclusive utilizing replace missing utility.
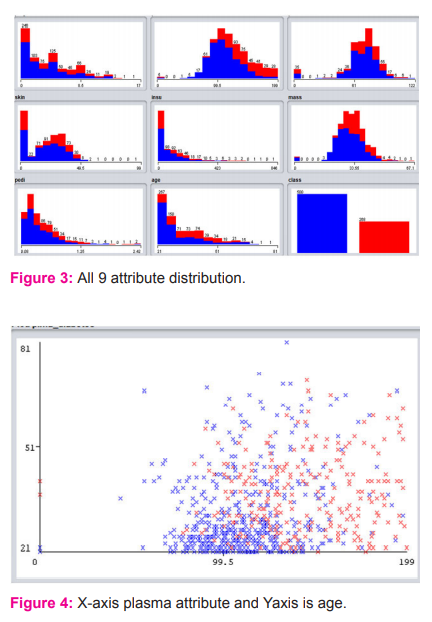
The 768 cases and 9 attributes of pima diabetes with 10 fold cross validation gives output weights for different classes as 0.09162112439990752 and 0.12059259597282575. Figure 3 shows the 768 samples and 9 features of pima diabetes with 10 fold cross validation gives output weights for different classes as 0.916 and 0.120 ,Unit center weights are 1.422, 3.868, 0.027, 0.453, 0.871, 1.43, 1.265, 1.146. Output weights for different cases are12.179, 1.895. Unitcenter as 0.025, 0.296, 0.570, 0.197, 0.179, 0.111, 0.123, 0.208. Bias weights for different classes as -3.790 and 3.691 and time grabed to build model: 0.23 seconds. The correctly classified instances using RBF network is 585 with accuracy 76.1719%. Incorrectly Classified Instances are 183 with 23.8281%, Kappa statistic is 0.4553, mean absolute error is 0.338, root mean squared error is 0.404, relative absolute error is 74.3694%, root relative squared error is 84.7695%. The confusion matrix is 430, 70 as tested negative and 113, 155 as b as tested positive. The distribution of all 9 attributes is given in figure 4.
TP Rate FP Rate Precision Recall F-Measure MCC ROC Area PRC Area Class
0.860 0.422 0.792 0.860 0.825 0.459 0.819 0.890 tested_negative
0.578 0.140 0.689 0.578 0.629 0.459 0.819 0.684 tested_positive
Weighted Avg. 0.762 0.323 0.756 0.762 0.756 0.459 0.819 0.818
CONCLUSION
The problem identified is the automatic classification of diabetics data and the solution using deep learning perceptron and SVM for the best is accomplished. MLP deep-learning classifier with 18 trainable parameters of correctly classified 595 Instances with a classification accuracy of 77.474 % and Incorrectly Classified Instances of 173 with 22.526% are obtained. The SVM classifier Correctly Classified Instances are 500 with 65.1042 % and Incorrectly Classified Instances are 268 with 34.8958%. The classification accuracy can further be increased by fusion of attributes and using advanced neural network classifiers. Deep learning perceptron classifier performs well with diabetes dataset and can be used for further automatic identification and detection systems. In future, the number of attributes needs to be increased and the number of training set also need to be increased with sequential classifier and deep neural networks to reduce false negatives.
Conflict of Interest: None
Source of Funding: None
References:
1. Han W, Shengqi Y, Zhangqin H, Jian H, Xiaoyi W. Type 2 diabetes mellitus prediction model based on data mining, Infor Med Unlocked 2018;10:100–107.
2. Aishwarya R. A Method for Classification Using Machine Learning Technique for Diabetes, Int J Engg Tech 2013;5(3):2903-2908.
3. Nahla HB, Andrew P. Bradley, and Mohamed Nabil H. BarakatIntelligible Support Vector Machines for Diagnosis of Diabetes Mellitus, IEEE Transactions On Inform Techn. Biomed. 2010;14( 4).
4. Naz H, Ahuza S. Deep learning approach for diabetes prediction using PIMA Indian dataset. J Diab Metab Dis April 2020;19(1):391-403.
5. Swapna GVinayakumar RSoman K. Diabetes, Diabetes detection using deep learning algorithms. ICT express 2018;4:243-246
6. El-Jerjawi NS, Abu-Naser SS. Diabetes prediction using artificial neural network. Int J Adv Sci Tech 2018;121:55–64.
7. Perveen S, Shahbaz M, Keshavjee K and Guergachi A, Metabolic Syndrome and Development of Diabetes Mellitus: Predictive Modeling Based on Machine Learning Techniques. IEEE Access 2019;7: 1365-1375.
8. Sajida PN, Muhammad S. Performance Analysis of Data Mining Classification Techniques to Predict Diabetes. Procedia Comp Sci 2016; (82):115-121.
9. Barakat N, Bradley A, Barakat MN. Intelligible Support Vector Machines for Diagnosis of Diabetes Mellitus. IEEE Transactions Info Tech Biomed 2010;14(4):1114-1120.
10. AIyer S, Jeyalatha R, Ronak S, Diagnosis Of Diabetes Using Classification Mining Techniques. Int J Data Mining Knowl Man Pro 2015;5(1):1-14.
|






 This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License