IJCRR - 2(4), April, 2010
Pages: 30-43
Print Article
Download XML Download PDF
ISOLATION AND CHARACTERIZATION OF A BACTERIOPHAGE SPECIFIC TO DRUG RESISTANT KLEBSIELLA PNEUMONIAE DR1
Author: Wanpen Thamniamton, Varaporn Boonsarn, Parichat Phumkhachorn, Pongsak Rattanachaikunsopon
Category: General Sciences
Abstract:A lytic bacteriophage isolated from sewage water was found to attack drugresistant Klebsiella pneumoniae DR1 isolated from a clinical specimen. The phage, designated kpdr1, had an isomeric head (60 nm in diameter) with a contractile tail (93 nm long and 13 nm wide). It was identified in Myoviridae family. From the host range test, the phage was highly specific to K. pneumoniae DR1. A onestep growth experiment of phage
kpdr1 at 37 C after the phage infection showed that the latent period was 30 min, the burst period was 180
min, and the average burst size was about 52 phage particles/infected cell. The phage was able to survive after incubating at 37 C for 24 h in pH 5-12. The phage was also stable at the temperature of 60 C for at least 3 min. Various disinfectants could inactivate the phage after treating at 37 C for 24 h. Knowledge of the properties of K. pneumoniae DR1 bacteriophage kpdr1 may be important for the development of controlled infection.
Keywords: Bacteriophage, Klebsiella pneumoniae, drug resistant bacteria, phage therapy
Full Text:
Introduction
Klebsiella pneumoniae is a gramnegative bacterium which clinically the most important member of the Klebsiella genus of Enterobacteriaceae. It causes urinary tract infection (UTI), pneumonia, and abdominal infection. It normally affects persons with immunocompromised such as hospital patients, diabetes patients and people with chronic lung disease. Usually, alcoholics also suffer from K. pneumoniae infection. Thus, the infections are either hospital-acquired or community-acquired[1]. Treatment for K. pneumoniae infection is usually done by using antibiotics such as cephalosporins, quinolones, or aminoglycosides. However, there is a widespread development of antibiotic resistant strains of K. pneumoniae[2], so the need for new antibiotics and alternative approaches to control drug-resistant K. pneumoniae infection is receiving increased attention[3]. One such alternative is the possible therapeutic use of bacteriophage or bacteriophage therapy[4,5]
Bacteriophages or phages are the viruses that infect bacteria. There are many of different phages, each of which may infect only one or several types of bacteria. Phages are common in all natural environments and are directly related to the numbers of bacteria present[6]. When a phage meets a suitable host bacterium, it binds to the particular molecules on the outermost part of the host and then injects its genetic material into the host cell. Lytic phages instruct the machinery in the host cell to make more phages. Fully viable progeny phages burst out and kill the bacteria. The released phages attack new bacteria. This process continues until all the bacteria are eliminated from the ecosystem[5] .
ystem[5] . One of the advantages of phage therapy over antibiotics is that phages infect only specific bacteria, so they can kill only the harmful bacteria without affecting the beneficial bacteria present in the body. On the other hand, antibiotics usually target both pathogenic and normal flora. Thus, phage therapy is considered safer for therapeutic use[5]. The application of phages to treat bacterial infections has been successfully used both in infected human and experimentally infected animal[7] . In this study, we isolated a phage, kpdr1, against drug-resistant K. pneumoniae DR1 from sewage water and also characterized the phage in some aspects. The phage from this study may be useful as a potential therapeutic agent for controlling K. pneumoniae DR1 infection which usually occurring in our communities.
Materials and Methods Bacterial Strains and Culture Conditions
K. pneumoniae strain was isolated from a patient hospitalized at the Sappasitiprasong Hospital, Ubon Ratchathani, Thailand and was designated as DR1 in our nomenclature. The strain was grown in Brain Heart Infusion (BHI) medium (HiMedia, Mumbai, India) at 37 C and confirmed as K. pneumoniae by the standard biochemical tests. Antibiotic susceptibility testing by Kirby-Bauer?s method revealed that the K. pneumoniae DR1 strain was resistant to most of the commonly used drugs. Staphylococcus aureus ATCC25923 and Escherichia coli ATCC25922 were used as control strains in antibiotic susceptibility testing.
To test the host range of phage, kpdr1, several bacterial strains were used (Table 1). All of the bacterial strains used in this study were grown in BHI medium at 37 C. The bacterial cultures were stored as stock cultures in BHI broth supplemented with 20% (v/v) glycerol at -70 C.
Phage Isolation
Sewage water samples for phage isolation were obtained from the sewage treatment tank located at the Sappasitiprasong Hospital. The samples were stored at 4 C overnight to allow larger suspended sediments to settle out. These crudely clarified samples were centrifuged (4,500 g for 10 min) to remove bacterial cells and debris. The supernatant was passed through a 0.22 m membrane filter (SartoriusAG, Goettingen, Germany). For phage enrichment, the filtrate was added to equal volume of double strength BHI broth and inoculated with an early log phase host culture, K. pneumoniae DR1. After incubation at 37 C overnight, the culture was centrifuged (4,500 g for 10 min) and the supernatant was passed through 0.22 m membrane filter. The filtrate was stored at 4 C and subjected to the following phage detection.
Phage Detection and Host Range Determination
Phage detection was performed by using a spot test method[8]. The test was used as an initial step for the presence of phage by observing lytic activity of phages. Soft agar in 5 ml (BHI broth with 0.7% agar) was seeded with 0.1 ml of a log-phase host culture, mixed thoroughly, and poured onto a BHI agar plate. After solidification, 10 l of phage filtrate was spotted onto the top agar layer. After drying, the plate was incubated at 37 C overnight. A clear zone in the plate, resulting from the lysis of host cells, indicated the presence of phage. Spot tests were also used for host range studies. The procedures were as described above but using the other bacterial hosts listed in the table 1.
Phage Propagation
Phage was purified by single plaque isolation[8] using K. pneumoniae DR1 as a propagating strain. A single plaque was picked from the lawn of the bacterial host, and propagated in 10 ml of an early log phase K. pneumoniae DR1 culture in BHI broth. After incubated at 37 C overnight, phage lysate was centrifuged at 4,500 g for 10 min. The supernatant was filtered through 0.22 m membrane filter. Phage stock was stored at 4 C, and an aliquot was frozen at -70 C. Phage titer was counted as plaque-forming unit (pfu/ml) using the double-layer agar plaque method[9] .
One-Step Growth Curve
For one-step growth experiments, a method of Capra et al.[10] was used. A mid-exponential phase culture of K. pneumoniae DR1 (optical density at 600nm = 0.5) was harvested and suspended in one-fifth of the initial volume of fresh BHI broth. Phages were added at a multiplicity of infection (MOI) of 0.5, and allowed to adsorb for 30 min at 37 C. Cells were harvested by centrifugation (10,000 g for 5 min), and resuspended in BHI broth. Decimal dilutions were made, incubated at 37 C, and at intervals, aliquots from each dilution were collected for phage counts. Latent period, burst time and burst size were calculated from the one-step growth curve.
Phage Stability
Resistance to physical and biocides was determined according to the methods described by Lu et al.[8] and Capra et al.[10] with minor modifications. Thermal inactivation was examined at 50, 60, 70, and 80 C. A tube containing sterile, deionized water was preheated to a desirable temperature. Phage solution was added into the tube at final concentration of 106 pfu/ml. After heating at intervals between 30 s to 3 min, phage samples were taken for phage counts. Results were expressed as the concentration (pfu/ml) of active viral particles and its log plotted against time. Phage stability was also examined at pH values ranging from 2 to 13, after incubation for 24 h at 37 C. Phages were added into each pH value at final concentration of 106 pfu/ml. The surviving phages were immediately counted and the results were expressed as the concentration (pfu/ml) of active viral particles and its log plotted against pH values.
Resistance to disinfectants was also determined using commercial ethanol (10, 50, 75, and 95% v/v), isopropanol (10, 50, 75 and 95% v/v), and sodium hypochlorite (200, 400 and 600 ppm). After treating at different concentrations of each disinfectant for 24 h at 37 C, the surviving phages were counted and the results were expressed as a percentage of the initial viral counts.
Phage Preparation for Electron Microscopy
Phage preparation for direct visualization by electron microscopy was carried out as described by Watanabe et al.[11] with some modifications. Briefly outlined, five hundred milliliters of BHI medium was inoculated with a bacterial host, K. pneumoniae DR1, and grown to an optical density at 600 nm of 0.5. The bacterial host was then infected with 5 ml of a phage suspension (106 pfu/ml) and incubated overnight at 37 C. After centrifugation at 4,500 g for 20 min, the supernatant was collected, and was centrifuged at 4 C with a 70.1Ti rotor at 28,500 rpm for 1 h in a Beckman L- 80 ultracentrifuge (Beckman, CA, USA). The resulting pellets were resuspended in 5 ml of phage buffer (20 mM Tris-HCl [pH 7.4], 100 mM NaCl, 10mM MgSO4). A purified phage was recovered after centrifugation (4,500 g for 20 min) and the supernatant was passed through a 0.22 m membrane filter. The purified phage was stored at 4 C for electron microscopy.
Electron microscopy
A drop of the purified phage suspension was applied to a carboncoated grid for 5 min, then removed with a pipette and immediately replaced with a solution of 2% (w/v) uranyl acetate. After 1 min, the liquid was removed with a filter paper. The grids were examined in a JEOL JEM- 1230 transmission electron microscope (JEOL, Tokyo, Japan) at 80 kV accelerating voltage.
Results
Isolation and Morphology of Phage
Five sewage water samples were collected for phage isolation. One sample was obtained a phage specific to the drug resistant K. pneumoniae DR1, designated kpdr1. The phage was found to produce small central clear plaques (average diameter, 1.0 mm) surrounding with a large halo (Fig. 1). The phage had an isometric head that was 60 nm in diameter and a long contractile tail (13 93 nm) under the electron microscope (Fig. 1). The virion had a baseplate at the distal end of its tail but did not show other additional structures such as collars or fibers. The phage was classified as a member of the Myoviridae family[6] .
Host Range Determination
The host range of phage kpdr1 was determined by testing the formation of clear zones at 37 C after overnight incubation on lawns of 17 bacterial strains. The results, summarized in Table 1, showed that the phage had a highly specific host range. The phage was able to infect only K. pneumoniae DR1 strain. All the tested strains were insensitive to this phage.
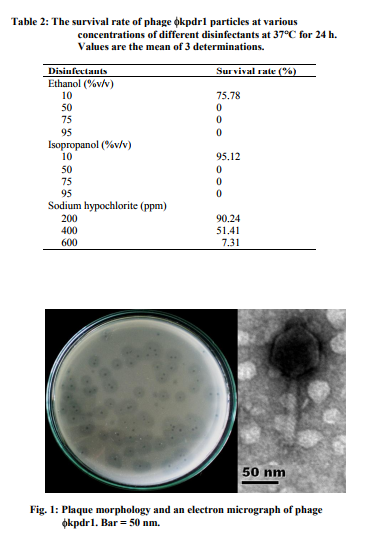
One-Step Growth Curve
Multiplication parameters of the lytic cycle of phage kpdr1 were determined from the one-step growth curve (Fig. 2). The latent and burst periods were 30 and 180 min, respectively, and the burst size was estimated at 52 phage particles per infected cell.
Phage Stability
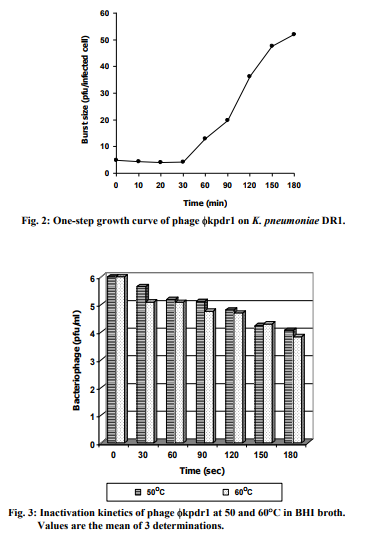
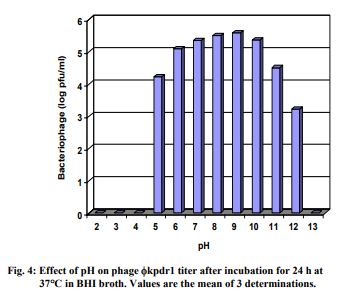
Phage kpdr1 was able to survive at 50 and 60 C. There was not much difference of phage counts between 50 and 60 C. Heating at 60 C for 180 s was insufficient for complete inactivation but was reduced the phage titer about 2 log (Fig. 3). However, the phage was totally inactivated at 70 and 80 C after only 30 s (data not shown). Phage kpdr1 maintained its infectivity when incubated at 37 C for 24 h in a pH range between 5 and 12 (Fig. 4). An obvious decrease in phage particle counts was observed at pH higher than 10 and at pH lower than 6. At pH 2, 3, 4, and 13, phage could not be detected. After phage kpdr1 suspensions were treated with various concentrations of different disinfectants at 37 C for 24 h, phage particles mostly became inactivated (Table 2).
Discussion
For more than half a century, the human society has been relying primarily on antibiotics to treat infection disease caused by pathogenic bacteria. However, the worldwide spread of pathogenic bacteria that are resistant to a variety of antibiotics has made antibiotics less and less effective[3]. Drug-resistant K. pneumoniae is a common nosocomial pathogen and a very important cause of morbidity and mortality in several countries[12-14]. This clearly highlights the need for new antibacterial agents with fundamentally different modes of action than that of traditional antibiotics to control drug-resistant K. pneumoniae infection. Bacteriophages or phages represent the new class of antibacterial agents. Obviously, lytic phages are the best candidates for bacteriophage therapy as they undergo rapid growth, disrupt bacterial metabolism and reproduction, and lyse the bacterial cells. In this study, we found a lytic phage, kpdr1, against drug-resistant K. pneumoniae DR1 in sewage water from the local hospital where the host strain was isolated. Not surprisingly, phages can be found in places where their hosts exist[9,15]. Because phage kpdr1 use K. pneumoniae DR1 as a host organism, it can be found in places where its host is plentiful.
Transmission electron micrograph showed that phage kpdr1 consisted of an isometric head and a long contractile tail. According to its morphology, this phage was classified as a member of the Myoviridae family. This virion morphology was similar to that of phage Kpp55 of K. pneumoniae which identified as the T-even like phage group[16]. However, the morphology of phage against K. pneumoniae found in this study was different from other previous reports such as phage K 1, KPO1K2, SS, and Kpn phage group all of which belonged to the Podoviridae family having an icosahedral head with a short noncontractile tail[17-19]. It is possible that further analysis of phage kpdr1 will indicate that it represents new virus family or similar to the previous families of viruses found in K. pneumoniae. The diversity of phage types against a single species is truly remarkable.
On the basis of spot test to check the host range, phage kpdr1 was found to infect only drug-resistant K. pneumoniae DR1 strain. None of the ther bacterial hosts used in the present study showed susceptibility to this phage. Interestingly, the phage was not even able to infect the two other strains of K. pneumoniae. Thus, the host range of phage kpdr1 seems to be extremely narrow. The narrow host range of phage should be advantageous, in principle, as phage therapy results in less harm to the normal flora than commonly used antibiotics. Moreover, the high specificity of phage and its host could be applicable for phage typing in epidemiological studies as well[20] .
Several studies documented that thermal and pH stability and chemical resistance of phages varied depending on types of phage[8,10,18]. Therefore, it is of interest to investigate the stability of phage kpdr1 in a wide range of temperature and pH and in some common used biocides. The results revealed that phage kpdr1 was quite stable in a broad pH range (5-12) and could be survived at 60 C for at least 3 min. However, phage kpdr1 was hardly able to survive in all disinfectants used in this experiment. This information may be useful for broad applications of the phage in phage therapy or in other applications.
In conclusion, this study finds a lytic phage, kpdr1, infecting a drugresistant K. pneumoniae DR1 strain that will lead to a better controlling infection especially in the outbreak area. Further research is needed to evaluate the potential therapeutic use of the phage to control the drug-resistant K. pneumoniae DR1 infection in animal models.
Acknowledgement
The authors gratefully acknowledge Nual-anong Narkong, the Instrumental Center, Faculty of Science, Mahasarakham University for her advice and assistance with electron microscopy.
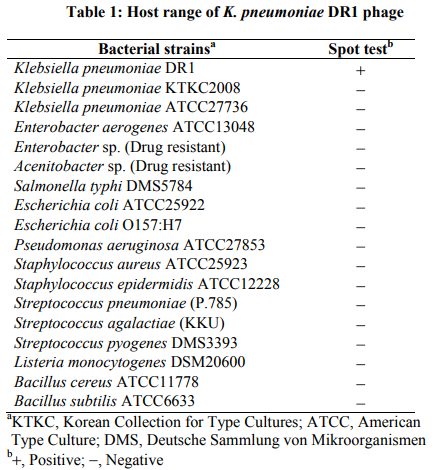
References:
1. Tsay RW, Siu LK, Fung CP, Chang FY. Characteristics of bacteremia between community-acquired and nosocomial Klebsiella pneumoniae infection: risk factor for mortality and the impact of capsular serotypes as a herald for community-acquired infection. Arch Intern Med 2002; 162: 1021- 27.
2. DeRyke CD, Pharm D, Wallace MR. Antimicrobial resistance update: Klebsiella pneumoniae carbapenemases. Drug Benefit Trends 2009; 21: 238-40.
3. Parisien A, Allain B, Zhang J, Mandeville R, Lan CQ. Novel alternatives to antibiotics: bacteriophages, bacterial cell wall hydrolases, and antimicrobial peptides. J Appl Microbiol 2008; 104: 1-13.
4. Sulakvelidze A, Alavidze Z, Morris JGJr. Bacteriophage therapy. Antimicrobe Agents Chemother 2001; 45: 649-59
. 5. Sandeep K. Bacteriophage precision drug against bacterial infections. Curr Sci 2006; 90: 631- 33.
6. Ackermann HW. Bacteriophage observations and evolution. Res Microbiol 2003; 154: 245-51.
7. Matsuzaki S, Rashel M, Uchiyama J, Sakurai S, Ujihara T, Kuroda M, et al. Bacteriophage therapy: a revitalized therapy against bacterial infectious disease. J Infect Chemother 2005; 11: 211-19.
8. Lu Z, Breidt JrF, Fleming HP, Altermann E, Klaenhammer TR. Isolation and characterization of a Lactobacillus plantarum bacteriophage, JL-1, from a cucumber fermentation. Int J Food Microbiol 2003; 84: 225-35.
9. McLaughlin MR, Balaa MF, Sims J, King R. Isolation of Salmonella bacteriophages from swine effluent lagoons. J Environ Qual 2006; 35: 522-8.
10. Capra ML, Quiberoni AL, Ackermann HW, Moineau S, Reinheimer JA. Characterization of a new virulent phage (MLC-A) of Lactobacillus paracasei. J Dairy Sci 2006; 89: 2414-23.
11. Watanabe K, Takesue S, Jin-Nai K, Yoshikawa T. Bacteriophage active against the lactic acid beverageproducing bacterium Lactobacillus casei. Appl Microbiol 1970; 20: 409-15.
12. Podschun R, Ullmann U. Klebsiella spp. As nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin Microbiol Rev 1998; 11: 589- 603.
13. Yan JJ, Ko WC, Tsai SH, Wu HM, Wu JJ. Outbreak of infection with multidrug-resistant Klebsiella pneumoniae carrying blaIMP-8 in a university medical center in Taiwan. J Clin Microbiol 2001; 39: 4433-9
. 14. Kang CI, Kim SH, Bang JW, Kim HB, Kim NJ, Kim EC, et al. Community-acquired versus nosocomial Klebsiella pneumoniae bacteremia: clinical features, treatment outcomes, and clinical implication of antimicrobial resistance. J Korean Med Sci 2006; 21: 816-22.
15. Sundar MM, Nagananda GS, Das A, Bhattacharya S, Suryan S. Isolation of host-specific bacteriophages from sewage against human pathogens. Asain J Biotechnol 2009, 1: 163-70.
16. Wu LT, Chang SY, Yen MR, Yang TC, Tseng YH. Characterization of extended-host-range pseudo-T-even bacteriophage Kpp95 isolated on Klebsiella pneumoniae. Appl Environ Microbiol 2007; 73: 2532- 40.
17. Malik R, Chhibber S. Protection with bacteriophage K 1 against fatal Klebsiella pneumoniae induced burn wound infection in mice. J Microbiol Immunol Infect 2009; 42: 134-40.
18. Verma V, Harjai K, Chhibber S. Characterization of a T7-like lytic bacteriophage of Klebsiella pneumoniae B5055: a potential therapeutic agent. Curr Microbiol 2009; 59: 274-81.
19. Kumari S., Harjai K, Chhibber S. Efficacy of bacteriophage treatment in murine burn wound infection induced by Klebsiella pneumoniae. J Microbiol Biotechnol 2009; 19: 622-8.
20. Hagens S, Loessner MJ. Application of bacteriophages for detection and control of foodborne pathogens. Appl Microbiol Biotechnol 2007; 76: 513-9
|






 This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License