IJCRR - 4(8), April, 2012
Pages: 33-42
Date of Publication: 25-Apr-2012
Print Article
Download XML Download PDF
DNA BARCODING, PHYLOGENETIC DIVERSITY STUDIES OF ETROPLUS SURATENSIS FISH FROM
POORANANKUPPAM BRACKISH WATER, PUDUCHERRY
Author: Sachithanandam V., Mohan P.M., Muruganandam N., Chaaithanya I.K., Arun Kumar P, Siva Sankar R
Category: General Sciences
Abstract:Etroplus suratensis is known for the high commercial value fish available in South India. The identification of the species of this fish cumbersome and inaccurate in different life stages of the fish. Therefore, DNA sequence of cytochrome Oxidase subunit I gene was analysed for the species identification and phylogenetic relationship of the species. The average genetic distance of conspecifics species value was found to be 0.005%. The present work suggests that COI sequence provides sufficient information on phylogenetic and evolutionary relationship to distinguish the Etroplus suratensis species, the brackish waters species of pearl spots, unambiguously. Further, this work revealed that every species having individual genetic distances depended upon the environmental stress and water quality, which play an important role for its minor morphometric variations. Therefore, it was concluded that a DNA COI barcoding tool can be used for fish identification by non technical personnel (other than taxonomist).
Keywords: DNA barcoding, COI, brackish water, Pooranankuppam and Etroplus suratensis
Full Text:
INTRODUCTION
The chromids or the pearl-spots (Family: Cichlidae) form an important group among the brackish water fishes of the tropics. One of the genus Etroplus contains E. suratensis fish is inhabitant in fresh water and brackish water in southern India. E. suratensis has many desirable features which make them ideal fishes for aquaculture like wide salinity tolerance, ability to breed in confined waters, fast rate of growth, good body weight, tasty flesh, highly adaptable feeding habits, robust, sturdy body1 . Experimental cultures of this species show its potential for polyculture and integrated farming with poultry. In addition to export, it has high demand in the local market and fetches a price of more than US$ 3/kg2 . These fish is available throughout the year. The average production is about 1000 kg/ha/year over 8-10 month growout period. Morphometric studies are not only essential to understand the taxonomy but also the health of a species (including reproduction) in an environment. The morphometric features of the fish are unique to the species whereas the variations in its feature are probably related to the habit and habitat3 . Morphometric measurements have been widely used to discriminate populations of various fish species4-6 . Fishes are considered to be phenotypically more variable than most other vertebrates, having relatively higher withinpopulation coefficients of variation of phenotypic characters. Genetic polymorphism or environmental factors may induce morphological variability among spatially separated fish populations7 , and phenotypic plasticity in fish morphology has been documented for various species, including cichlids8,9 . E. suratensis is known to have variations in various morphological features which are dependent on the geographical partition. Further environmental comparisons of these estuaries would be worthwhile in understanding the evolution of such variations. In addition, genetic investigations of the variation and differentiation involving more estuarine samples of E. suratensis will be useful in substantiating the conclusions. The genotypic and phenotypic variation of species is a prerequisite in conserving them. DNA barcoding is highly efficient method in the analysis of genetic divergences among species as well as for intra species-level identifications10 . Among the marine living organisms of the IndoWest Pacific, Teleosts are among the bestdescribed, even though their systematics and taxonomy still need considerable research effort11,12. Southeast Asia has been identified as one of the world‘s biodiversity hotspots based on both plant and animal diversity13 . Many time taxonomic ambiguities exist due to morphological and meristic similarities. Modern taxonomic work includes analysis of a host of other traits, including anatomy, physiology, behaviour, genes, and geography, yet morphological traits remain cornerstone14 . In such circumstances, DNA barcodes has revealed that this could be helpful even for larval stage fish taxonomical identification15. To facilitate DNA barcode identification of fishes, regional working groups are conjoining under the Fish Barcode of Life (FISH-BOL) initiative16, which seeks to establish a barcode reference sequence library for all fishes17. The phylogenetic systems, in combination with conservation genetics, provide a critical frame work for understanding diversity18 and predict vulnerability to exploitation of tropical reef fishes19 . As the morphometric measurements could lead to misidentification of the species in different life stages of fish especially E. suratensis, which would affect the conservation strategy and the market value of the same. Molecular taxonomy appears to be the best tool for the species identification and advantageous over the other method of taxonomy, so the effort was taken to identify the E. suratensis through molecular taxonomy.
METHODS
Study Area, Sample collection and preservation: Fishes were collected from local fish landings at Pondicherry brackish area of Pooranankuppam (Fig.1). The identification of fishes was done as described in FAO14. A piece of muscle in the lateral line was collected and stored in 95% ethyl alcohol at -20 C for DNA extraction. The DNA Isolation and PCR condition work was carried out in Department of Ocean studies and Marine Biology centre at Port Blair, Andaman and Nicobar Islands. Molecular Taxonomy: Total DNA extracted from 0.25g of muscle by the standard proteinase-K/ phenol-chloroformisoamyl alcohol-ethanol method20. PCR amplification of a 650bp DNA fragment coding COI gene of the mitochondrial (mt) DNA genome was amplified using published primer set11 . PCR components and conditions for 50 l reaction were as described in our previous work21. The PCR product was resolved in 1% Agarose gel and visualized using Gel Doc System, to confirm the presence of amplified product sized 650 bp. Nucleotide sequencing was performed using BigDye Terminator Cycle Sequencing kit, following manufacturer‘s instructions (Applied Bio-systems, Foster City, CA, USA).
Sequence Analysis: The DNA sequences of phenotypically identified fishes were assembled using the SeqMan II version 5.03 (DNASTAR). The sequence of Etroplus suratensis reference sequences retrieved from the NCBI GenBank were aligned using Clustal W pair-wise and multiple alignment of MEGA version 4.122 . Sequence divergence was calculated using the Kimura 2- parameter (K2P) model23 and the mid-point rooted Neighbour-joining (NJ) tree of K2P distances was created to provide a graphic representation of the species divergence24 (Fig. 2). RESULTS DNA barcoding is a unique concept with many innovative attributes undertakes continuous improvement in taxonomy. The estimated Etroplus suratensis species DNA sequences were submitted to GenBank under the mentioned accession number in Table 1. NCBI BLASTn result revealed that 21 reference sequences were matched with maximum identity of Etroplus suratensis of Puducherry as described in Table 2. The average genetic distance within the species (K2P) is 0.005. The species genetic evolutionary pair wise distance proximity was calculated by the species similarity of genetic base pair. The Etroplus suratensis DOSMB species closely related to the species (FJ237544 – 0.006: GU5666028 – 0.006: FJ347966 – 0.006 and AY263870 – 0.009) in the Indian waters and USA respectively (Table 3). The nucleotide composition of Etroplus suratensis from the present studied species is A = 23.4%, T = 29.9%, G = 18.6% and C = 28.1%. The average nucleotide composition of E. suratensis among the species level is noted as A = 23.48%, T = 30.08%, G = 18.1% and C = 28.36% (Table 4, Fig.3). The NJ and K2P genetic distances were created to provide a graphic representation of the patterns of divergences. Two distinct clad with two sub clad of the same species were recognized with more than 90% bootstrap value. These two sub clad formation was identified based on the independent assemblages of close related species with differences in region and environmental closeness. The results clearly shows that every species having individual genetic distances depended upon survival of any species adaptation of the environmental stress and its water quality, which play an important role of significant values of the genetic distances internally and morphometric minor variations externally. DISCUSSION Fishes are largest group of vertebrates, which exhibits remarkable diversity of morphological attributes and biological adaptations25. In these circumstances fish taxonomist facing a large problems while the identification of fishes. To overcome this problem, a morphology- based identification combined with molecular based approach for the species identification using DNA barcoding would be an ideal tool26 . This tool is an efficient method for species-level identification of the mitochondrial Cytochrome c Oxidase I (COI) gene27. Mitochondrial DNA (mtDNA) has been widely employed in phylogenetic studies of animals because it evolves much more rapidly than nuclear DNA, resulting in the accumulation of differences between closely related species 28-30 . This study provides the interspecific heterogeneity which enhanced the efficiency of species identification through bar coding. This was proved in Australian marine fishes11 , freshwater fish barcoding from Canadian31 and carangid fishes from Indian waters32 . The variations of phenotypic character of species are unique and it is probably related to the habit and habitat among the variants of this species33 . Genetic variation of the green chromid has not been studied previously of the genus E. suratensis in Indian waters. In the studies of bar coding, it has been reported that K2P values between two species should be greater than 0.0227,34 and in e.g. Indian mosquito DNA barcoding average K2P values is 0.032935 . The present bar code exhibited K2P pair wise genetic distances variation among the species level is 0.005. However, such variation has greater impact on the survival of the haplotypes and its evolution. The efficiency of species identification by molecular method was enhanced by the interspecific heterogenetic relationship displayed36 . Based on the above investigation it is clearly evident that the E. suratensis identified in Puducherry waters represented the haplotypes species morphometrically, the other part of the India and USA waters. However, bar code results suggested that they are genetically varied. Since, this species identified as haplotypes it may be of low level differences in morphometrically because of low genetic distances. Results of phylogenetic and evolutionary relationship of present study were supported by earlier studies11. Further, it has also confirmed that the DNA barcoding help to recover phylogenetic information and to understand the relationship with the species as well as Order level36. The present study also supported that mtDNA COI barcode region offers best species identification, which is most applicable and comparable with the mtDNA 12S - 16S rRNA region sequences37 . CONCLUSION The study concludes that the fish which identified morphometrically and DNA barcoding method using COI gene sequence are one and same species of E. suratensis, the brackish waters species of pearl spots. Further, this result also informed that every species having individual genetic distances depended upon the environmental stress and water quality which play an important role for its minor morphometric variations. Moreover, the mtDNA COI gene based identification provides high resolution in species identification in fishes.
ACKNOWLEDGMENTS
The Authors express their sincere acknowledges to Prof. J. A. K. Tareen, Vice-Chancellor of Pondicherry University and the constant help and encouragement of Dr. P. Vijayachari, Director, Regional Medical Research Centre (ICMR), Port Blair for the extension of facility during this study. The manuscript valuable review and commands supported by Chandal Lal and Sayi Dev. We express thanks to the Pondicherry University and Central Marine Living Resources and Ecology (CMLRE) for funding this work. Authors acknowledge the immense help received from the scholars whose articles are cited and included in references of this manuscript. The authors are also grateful to authors/ editors/ publishers of all those articles, journals and books from where the literature for this article has been reviewed and discussed.
References:
1. Hora, S. L and PilIay, T. V. R. (1962). Hand·book on the fish culture in the Indo·Pacific region. FAO Fish. BioI. Tech. pp. 14:124.
2. James, C. M. (2000). Potential of marine fish farming in India: www\Grp\Grouper\Research\Economics\20 00\2103.htm pp. 1-3.
3. Mauro José Cavalcanti., Leandro Rabello Monteiro and Paulo Roberto Duarte Lopes. (1999). Landmarkbased Morphometric Analysis in Selected Species of Serranid Fishes (Perciformes: Teleostei) Zool.stud. 38: pp. 287294.
4. Elliott, N. G., Haskard, K. and Koslow J. A. (1995). Morphometric analysis of orange roughy (Haplostethus atlanticus) off the continental slope of southern Australia. Journal of Fish Biology 46, 202-220.
5. Uiblein, F. (1995). Morphological variability between populations of Neobythites stefanovi (Pisces: Ophidiidae) from deep Red Sea and Gulf of Aden. Marine Ecology Progress Series 124: 23-29.
6. Hurlbut, T. and Clay, D. (1998). Morphometric and meristic differences between shallow and deep-water populations of whitehake (Urophycis tenuis) in the southern Gulf of St. Lawrence. Canadian Journal of Fisheries and Aquatic Sciences 55: 2274-2282.
7. Carvalho, G. R. (1993). Evolutionary aspects of fish distribution: genetic variability and adaptation. Journal of Fish Biology 43 (Supl. A), 53-73.
8. Wimberger, P. H. (1991). Plasticity of jaw and skull morphology in the neotropical cichlids Geophagus brasiliensis and G. steindachneri. Evolution 45, 1545-1561.
9. Wimberger, P. H. (1992). Plasticity of fish body shape. The effects of diet, development, family and age in two species of Geophagus (Pisces: Cichlidae). Biological Journal of the Linnaean Society 45, 197-218.
10. Suneetha Gunawickrama, K.B. (2007). Morphological heterogeneity and population differentitation in the green chromid Etroplus suratensis (pisces: Cichlidae) in Sri Lanka, Ruhuna journal of Science, 2: 70-81.
11. Ward, R. D., Zemlak, T. S., Innes, B. H., Last, P. R. and Hebert, P. D. N. (2005). DNA barcoding Australia‘s fish species, Philos. Trans. R. Soc. B., 360: 1847–1857.
12. Knowlton, N. (2000). Molecular genetic analyses of species boundaries in the sea, Hydrobiologi 420: 73–90.
13. Myers, N., Mittermeier, R. A., Mittermeier, C. G., Fonseca G. A. B. and Kent, J. (2000). Biodiversity hotspots for conservation priorities. Nature 403: 853–858.
14. Fish Base. (2004). FishBase. World Wide Web electronic publication www. Fishbase.org.
15. Packer, L., Gibbs, J., Sheffield, C. and Hanner, R. (2009). DNA barcoding and the mediocrity of morphology. Molecular Ecology Resources, 9: 42–50.
16. Swartz, E. R., Mwale, M. and Hanner, R. (2008). A role for barcoding in the study of African fish diversity and conservation. South African Journal of Science, 104: 293– 298.
17. Ward, R. D., Hanner, R. H. and Hebert, P. D. N. (2009). The campaign to DNA barcode all fishes, FISH-BOL. Journal of Fish Biology, 74: 329–356.
18. Jean-Pierre Fe´ral. (2002). How useful are the genetic markers in attempts to understand and manage marine biodiversity? Journal of Experimental Marine Biology and Ecology, 268: 121– 145.
19. Simon Jennings., John, D. R., Nicholas, V. C. and Polunin. (1999). Predicting the Vulnerability of Tropical Reef Fishes to Exploitation with Phylogenies and Life Histories. Conservation Biology., 13: 1466- 1475.
20. Sambrook, J., Fritsch E. F. and Maniatus, T. (1989). Molecular Cloning: A Laboratory Manual, second edition. Cold spring Harbor Laboratory press, Cold Spring harbour, New York.
21. Sachithanandam, V., Mohan, P. M., Dhivya, P., Muruganandam, N., Baskaran, R., Chaaithanya, I. K. and Vijayachari, P. (2011). DNA barcoding, phylogenetic relationships and speciation of Genus: Plectropomus in Andaman coast Journal of research in Biology 3: 179-183.
22. Tamura, K., Dudley, J., Nei, M. and Kumar, S. (2007). MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Molecular Biology and Evolution
24: 1596-1599. 23. Kimura. (1980). A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. Journal of Molecular Evolution 15: 111–120.
24. Saitou, N. and Nei, M. (1987). The neighbour-joining method: A new method for reconstructing phylogenetic trees. Molecular Biology and Evolution 4: 406- 425.
25. Nelson, J. S. (2006). Fishes of the World, fourth ed. John Wiley & Sons, Hoboken, NJ.
26. Steinke, D., Zemlak, T. S., Boutillier, J. A. and Hebert, P. D. N. (2009). DNA barcoding of Pacific Canada‘s fishes. Mar. Bio 156: 2641–2647.
27. Hebert, P. D. N., Ratnasingham, S. and DeWaard, J. R. (2003a). Barcoding animal life: Cytochrome c oxidase subunit 1 divergences among closely related species. Proc R Soc Lond B Biol Sci 270: S596– S599.
28. Brown, W. M., George, M. and Wilson, A. C. (1979). Rapid evolution of animal mitochondrial DNA. Proc Natl Acad Sci USA 76: 1967–1971.
29. Moore, W. S. (1995). Inferring phylogenies from mtDNA variation: Mitochondrialgene trees versus nuclear-gene trees. Evolution, 49: 718–726.
30. Mindell, D. P., Sorenson, M. D., Huddleston, C. J., Miranda, H. C. and Knight, A., et al. (1997). Phylogenetic relationships among and within select avian orders based on mitochondrial DNA. In: DP Mindell, editor. Avian molecular evolution and systematics, New York: Academic Press. pp. 214–247.
31. Hubert, N., Hanner, R., Holm, E., Mandrak, N.E., Taylor, E., Burridge, M., Watkinson, D., Dumont, P., Curry, A., Bentzen, P., Zhang, J., April, J. and Bernatchez, L. (2008). Identifying Canadian freshwater fishes through DNA barcodes. PLos ONE, 3: 2490–2490.
32. Persis, M., Reddy, A. C. S., Rao, L. M., Khedkar, G. D., Ravinder, K. and Nasruddin, K. (2009). COI (cytochrome oxidase-I) sequence based studies of Carangid fishes from Kakinada coast, India. Molecular Biology Report. 36: 1733-1740.
33. Manimegalai, M., Karthikeyeni, S., Vasanth, S., Arul, G. S., Siva, V. T. and Subramanian, P. (2010). Morphometric Analysis – A tool to identify the different variant in a fish species E. Maculatus. International journal of Environmental Sciences, 1: 1-17.
34. Hebert, P. D. N., Cywinska, A., Ball, S. L. and DeWaard, J. R. (2003b). Biological identifications through DNA barcodes. Proc R Soc Lond B Biol Sci 270: 313–321.
35. Kumar, N. P., Rajavel, A. R., Natarajan R. and Jambulingam, P. (2007). Morphology, Systematics, Evolution DNA barcodes can distinguish species of Indian Mosquitoes (Diptera: Culicidae). Journal of medical Entomol 44: 1-7
36. Lievens, S., Goormatching, S. and Holsters, M. (2001). A critical evaluation of differential display as a tool to identify genes involved in legume nodulation: looking back and looking forwared. Nucl. Acids Res, 29: 3459 – 3468.
37. Ward, R. D., Holmes, B. H., White, W. T. and Last, P. R. (2008). DNA barcoding Australian chondrichthyans results and potential uses in conservation. Marine and Freshwater Research 59: 57-71
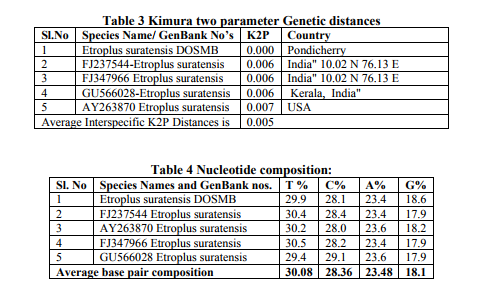
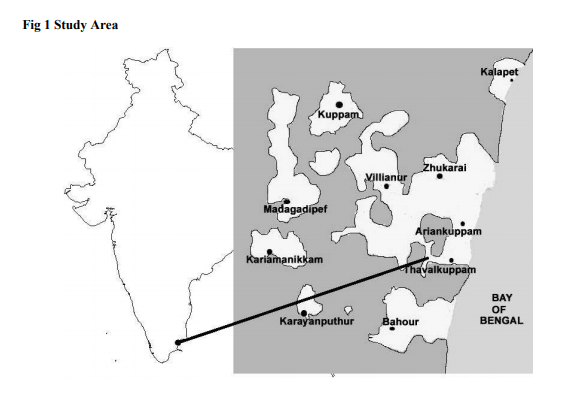
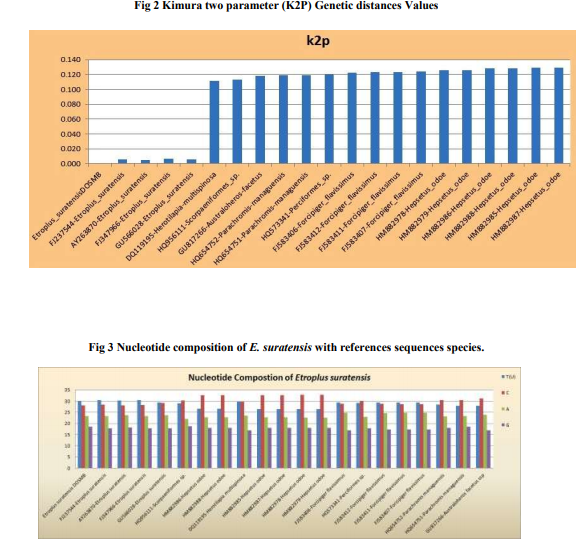
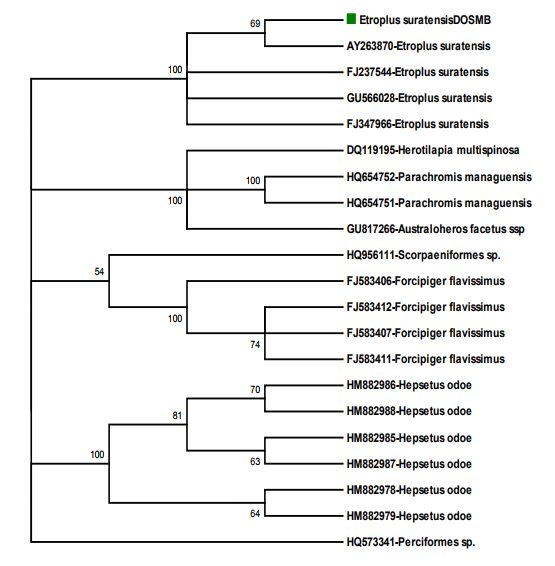
Fig 4 Neighbour-Joining (NJ) Method for Phylogenetic analysis and Evolutionary relationships of E. suratensis with NCBI references sequence The bootstrap test (1000 replicates) is shown next to the branches length is = 0.41765812 [Felsenstein 1985]. The evolutionary distances were computed using the Maximum Composite Likelihood method. Codon positions included were 1st+2nd+3rd. Phylogenetic analyses were conducted in MEGA4 [Tamura, et al 2007].
 Method for Phylogenetic analysis and Evolutionary relationships of E. suratensis with NCBI references sequence The bootstrap test (1000 replicates) is shown next to the branches length is = 0.41765812 [Felsenstein 1985]. The evolutionary distances were computed using the Maximum Composite Likelihood method. Codon positions included were 1st+2nd+3rd. Phylogenetic analyses were conducted in MEGA4 [Tamura, et al 2007]. )
|






 This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License