IJCRR - 4(12), June, 2012
Pages: 154-162
Date of Publication: 22-Jun-2012
Print Article
Download XML Download PDF
BLOOD STREAM INFECTIONS IN INTENSIVE CARE UNIT PATIENTS: A SINGLE CENTRE RETROSPECTIVE STUDY OF DISTRIBUTION AND ANTIBIOTIC RESISTANCE PATTERN IN CLINICAL ISOLATES
Author: Anamika Vyas, Ramavtar Saini, Pooja Gangrade, Mrityunjay Kumar
Category: Healthcare
Abstract:Introduction: Blood stream infections are an important cause of serious morbidity and mortality. Blood stream infections occur more frequently in patients hospitalized in intensive care units than in other units.
The knowledge of distribution and antibiotic resistance pattern of causative microbial agents in these infections is important for their prevention and empirical antibiotic treatment while awaiting culture and sensitivity results. Aim: The aim of the study was to describe the distribution of etiological agents causing blood stream infection in intensive care unit patients and their antibiotic resistance pattern, so that a guideline can be formulated for clinicians to choose an effective empiric antibiotic therapy. Methods: This was a retrospective study. Samples representing blood stream infections were sent from ICU to icrobiology laboratory for culture and sensitivity testing. Microbiological profile and antibiotic resistance pattern of clinical isolates was studied. Results: Positive cultures were obtained in 42 (11.9%) cases. Among culture positive isolates 56.1% were gram positive bacteria wherease (39%) and (4.9%) were gram negative bacilli and yeast respectively. Coagulase negative staphylococci (31.7%) was predominant organism followed by E.coli (24.4%), Staph aureus (12.2%), Enterococcus faecium (9.8%),Klebsiella pneumonia(7.3%) and nonfermenters (7.3%), Linezolid, Quinpristin/Dalfopristin, Vancomycim was the most active drugs for gram positive cocci wherease Amikacin and Carbapenams were most active drugs for gram negative bacilli. Nonfermenters showed multidrug resistance and were sensitive to tigecycline and colistin. Conclusion: This study provide information on distribution and antibiotic resistance pattern of microorganism causing blood stream infection. It may be a useful guide for physician to start empiric antibiotic therapy in cases of blood stream infections and in formulating antibiotic therapy policy in our hospital. It will provide a baseline reference data for future studies to detect changes in distribution and antibiotic resistance pattern of clinical isolates causing blood stream infection.
Keywords: Blood stream infection (BSI), Intensive Care Unit (ICU), Antibiotic resistance, Gram positive, Gram negative.
Full Text:
INTRODUCTION
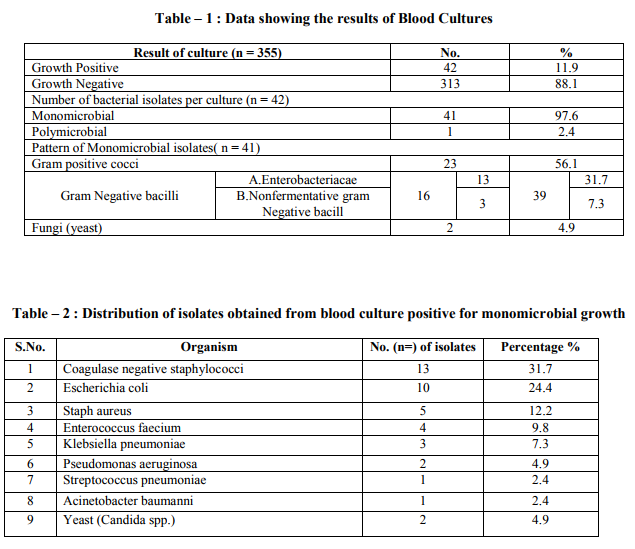
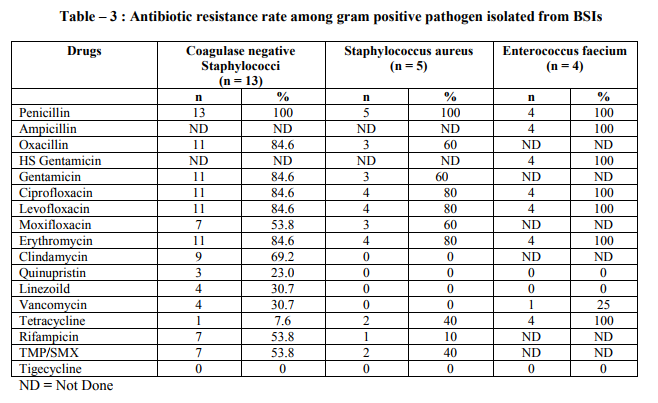
Blood stream infections (BSIs) are an important cause of serious morbidity and mortality and are among the most common health care associated infection.1 Illness associated with blood stream infection ranges from self limiting infections to life threatening sepsis that require rapid and aggressive antimicrobial treatment.2 Blood stream infections (BSIs) occur more frequently in patients hospitalized in intensive care units than in other units. Debilitated condition of the patient due to underlying diseases, invasive diagnostic and therapeutic procedures, contaminated life support equipment, prolonged use of in-situ invasive devices, therapy with multiple antimicrobials predispose these patients to life threatening BSI.3,4 BSIs in such patients not only extends the hospital stay but also causes an increase in hospital mortality rate and cost of care. Throughout the 1960s and 1970s, gram negative organisms were most frequently isolated from patients with nosocomial BSIs. Since then, infections due to gram positive organisms have become increasingly frequent.5,6,7 With the spread of multidrug resistant bacteria, the treatment of BSI has become a challenging task for the Physician when selecting a regimen with which to treat patients because in almost all cases, antimicrobial therapy is initiated empirically before the results of blood culture are available. This is only possible with knowledge of the most frequently isolated etiological agents and their likely antimicrobial resistance pattern in a given place. Early initiation of appropriate antimicrobial treatment is critical in decreasing morbidity and mortality among patients with blood stream infections.8 Keeping in mind that the profile of BSIs varies between institutions and the high mortality and morbidity associated with disease, a right choice of empiric therapy is of utmost importance. Therefore, the present retrospective analysis was carried out to determine the distribution of microbial agents responsible for bloodstream infections in ICU of a tertiary care hospital and to get an up dated knowledge about their resistance pattern. This will not only help the clinician in selecting the antibiotics for empirical therapy till the result of culture and sensitivity are known but also help the clinician in identifying the changing pattern of etiological agents and their drug resistance pattern for future reference as well as in formulation of guideline on antibiotic prescribing policy and other infection control measures. MATERIALS AND METHODS Present study was based on the retrospective analysis of data about blood culture results of specimen submitted for culture to Microbiology laboratory. Permission was taken from Institutional Human Research Ethics Committee for the study. A total of 355 blood samples were processed during study period of 6 months (Oct. 2011 to march 2012). All the samples were collected from intensive care units (Medicine, Surgery, Paediatrics) of GMCH – a 750 bedded, tertiary care, teaching hospital providing a full range of medical, surgical and super specialty facilities. Processing of samples was done at the department of microbiology. The patients hospitalized more than 48 hours in the ICU were included in this study. The diagnosis of BSI was based on criteria of the center for disease control.9 Patients presenting with the evidence of infection at the time of admission and patients with a hospitalization period of less than 48 hours in ICU were excluded from study.Under the appropriate aseptic precautions 8-10ml of blood from adults and 1-3ml of blood from children was drawn by venipuncture and inoculated into BACT/ALERT FA/PF disposable blood culture bottle commercially available (by BIOMERIEUX,FRANCE). After inoculation blood culture bottles were sent to microbiology lab where the inoculated bottles were vented in BACT/ALERT 3D a fully automated blood culturing system by (BIO MERIEUX FRANCE). Anaerobic blood culture media was not used. Positive growth was identified up to species level by colonial morphology, gram staining and biochemical reactions utilizing automated identification system (VITEK2 COMPACT SYSTEM, BIOMERIEUX, FRANCE) according to manufacturer‘s instructions. Isolation and identification of bacteria was followed by susceptibility testing that was performed with(VITEK2 COMPACT SYSTEM,BIOMERIEUX, FRANCE), applying the criteria suggested by the clinical and laboratory standard institute.10 Any result from the same patient with the same organism identification and the same sensitivity pattern received with in five days was considered a repeat culture and is counted only once in data base. An analysis of the distribution and the antibiotic resistance pattern of bacterial isolates was performed. RESULT During the six months study period 355 samples from patients admitted in ICU‘s of our tertiary care hospital were analyzed retrospectively. 42 (11.9%) blood culture samples were positive for growth. Of these 41 (97.6%) were monomicrobial and 1 (2.4%) was polymicrobial. Among 41 monomicrobial growth, 39 (95.1%) yielded growth of bacterial isolates and 2 (4.9%) yielded growth of yeast (candida). Among bacterial isolates 23 (56.1%) were gram positive cocci and 16 (39%) were gram negative bacilli (Table-1) The most common microorganism isolated from blood culture sample was Coagulase negative staphylococci (CoNS) (31.7%) followed by Escherichia coli (24.4%), Staph aureus (12.2%), Enterococcus faecium (9.8%), Klebsiella pneumoniae (7.3%), Nonfermenters (7.3%) (Table-2). The antibiotic resistance pattern of gram positive and gram negative organism is shown in the (Table-3 and 4) respectively. Oxacilln resistance was noted in 84.5% and 60% isolated strains of Coagulase negative staphylococcci and Staph aureus respectively. 8 out of 13 isolated strains of Enterobacteriacea (61.53%) were positive for ESBL test. Antibiotic resistance among Nonfermenter : Pseudomonas aeruginosa and Acinetobacter baumanii complex strains isolated displayed multidrug resistance (MDR-P. aeruginosa and Acinetobacter were defined as resistant to three or all four following antibiotics: (Ceftazidime, Ciprofloxacin, Gentamicin and Imipenam). They were sensitive to Colistin and Tigecycline respectively. (Data not shown as the number of isolates were very low). DISCUSSION Patient with blood stream infections have remained a challenge for clinicians to treat. Prompt diagnosis and effective treatment are necessary to prevent complications and to reduce mortality from BSI. Data from Ibrahim et.al11showed that mortality rates doubled up from 30% to 60% when inappropriate empirical therapy was given to ICU patients with BSI. Knowledge of the hospital epidemiology and antimicrobial susceptibility pattern of blood isolates help physician to effectively manage blood stream infections because considerable differences in the distribution and antibiotic resistance of blood isolates are reported even from hospital of similar size and mixture of patients of the same country. On the basis of prior knowledge of common causative agents and their susceptibility to prescribed antibiotic ,empiric therapy is started and later changed according to final culture and susceptibility report. In the present retrospective analysis 11.9% cultures were positive for growth. It has been reported between 11.6%-42% in different studies.12-17This variation probably reflects different populations, clinical settings, age groups, selection of patients, number of blood cultures collected, blood culture medium formulation, type of blood culture system used for bacterial detection.
Most of the cultures (97.6%) in the present study yield mono microbial growth. The poly microbial growth isolation rate was 2.4%. The reported polymicrobial isolation rate varies between 1 to 15 percent. The polymicrobial growth could mean contamination or a severe infection with bad prognosis.18,19 In our study gram positive bacterial isolates were more (56.1%) than gram negative bacilli (39%).This finding is in accordance with the studies by Falagas et al.12,Arora et al.14 , Karlowsky et al.20and Ahmed et al.21Among the gram positive bacterial isolates, Coagulase negative staphylococci (CoNS) accounted for (31.7%) wherease S. aureus was isolated in 12.2%cases. Our results are consistent with the result of other studies. Data from the SCOPE (Surveillance and control of pathogen of epidemiologic importance) project revealed that the most common pathogen causing nosocomial BSI were CoNS (32%) and S.aureus (16%).6 Another data published from national nosocomial surveillance system for ICU associated primary blood stream infections identified CoNS (37%) and S. aureus (12.6%) as the leading pathogen. The increase in the frequency of CoNS BSI isolates can be explained by increased use of invasive intra vascular devices. The trend for CoNS may reflect a change from regarding these organism as skin flora to viewing them as clinically significant. The interpretation of blood cultures positive for Coagulase negative staphylococcci has inherent difficulties and require careful reasoning. Gram negative pathogens were lower on the rank of organism and included E.coli (24.4%) followed by Klebsiella pneumoniae (7.3%) and Pseudomonas aeruginosa (4.9%). These results are similar to other study where these organisms have been among the leading gram negative pathogen.22 Candida were isolated in (4.9%) cases. This is consistent with the study of Arora et al.14 and Narain et al.23 where as in other studies the incidence is much higher.24 Extensive use of antibiotics, aggressive treatment of neoplastic diseases, an expanding population of patients with AIDS with prolonged survivors, use of indwelling devices for ICU and many other factors are responsible for considerable prevalence of fungemia. Increased antimicrobial resistance rate among microorganisms isolated from BSI are a significant problem worldwide. In developing country like India, although the majority of the population depends on public health care system, increasing fraction is being managed by private facilities. Heterogenecity is also reflected in health care practices. As a result different patterns of antimicrobial resistance and antimicrobial use may emerge with in country. Methicillin resistant Staph aureus (MRSA), Vancomycin resistant enterococi (VRE), Extended spectrum β-Lactamase producing Klebsiella sp. and E-coli, Carbapenam resistant enterobacteriaceae,pseudomonas aeruginosa,acinetobacter spp. are seen more frequently in ICU patient than in non-ICU patients in many countries. In the present study 60% of the S. aureus and 84.5% of the Coagulase negative staphylococci isolated showed resistance to oxacillin. our findings coincides with the studies by Falagas et al.12, Karlowsky et al.20 who also reported considerable proportion of S. aureus and CoNS resistant to oxacillin. With other tested antibiotic the rate of resistant was reasonably high. However we observed that no strain of S.aureus isolated showed resistance to vancomycin. Similar finding was noted in other studies.12,15 So vancomycin can be safely used in multidrug resistant strain. Linezolid and quinpristin/ dalfopristin , tigecycline were sensitive in all isolated strain of S.aureus . Rifampicin and clindamycin had a good activity against S.aureus so these drugs can be used as a cost effective alternative for S. aureus treatment as they are less expensive. Enterococi displayed markedly high level of drug resistance to most commonly used antibiotics. All the enterococcal isolates in present study were resistant to ampicillin, quinolones, high strength Gentamicin. Resistance to vancomycin was noted in 1 isolate .(25%). No resistance was seen against linezolid and quinpristin/dalfopristin. In another study 50- 60% enterocci isolates were resistant to all antibioties tested.25 In present study gram negative bacilli showed high resistance rate to majority of antibiotics. Other workers also have reported majority of gram negative isolates in their study as multidrug resistant.16 Ampicillin/sulbactam, ceftriaxone and other cephalosporins, quinolones (ciprofloxacin) showed high resistance for gram negative bacteria. These drugs have been commonly over used in out patients for many years hence high resistance rate is expected. The alarming finding in the present study was higher rate of resistance in enterobacteriaceae to cephalosporins which is a marker for presence of ESBL. In present study 61.5% isolated strain of enterobacteriaceae showed positive ESBL test. In India ESBL production among enterobacteriaceae has been reported to be between 74.4 %- 80.9%. This high rate of resistance to cephalosporin is due to abundant use of cephalosporins in the hospitals. It is a very trouble some development as the mortality is reported much higher with ESBL producing enterobacteriaeae. High resistance rate of gram negative bacilli can also be explained by the site where our study was performed (ICU‘s only) and the higher percentage of extended spectrum β-lactamases among gram negative bacilli (61.5%) which limits the therapeutic options in infection caused by such strain due to two broad factors: cross resistance (eg. to aminoglycosides, cotrimoxazole or fluoroquinolones) and the spectrum of these enzymes. In all vancomycin, linezolid and quinpristin/dalfopristin were most effective drugs for gram positive pathogens whereas amikacin and carbapenams were most active for gram negative bacilli(Enterobacteriaceae). Although the number of nonfermenters isolated in present study was less but the alarming finding was that isolated nonfermenters were multi drug resistant sensitive to only tigecycline and colistin (Data not shown). Doripenam and tigecycline are now available but with the degree of resistance encountered it is a matter of time before these antibiotics are exhausted. CONCLUSION The present retrospective analysis provided much needed information on the distribution of bacterial pathogens in blood stream infections and their antibiotic resistance pattern. The study conducted showed both Gram positive and Gram negative bacteria were responsible for blood stream infections. We observed resistance to several antimicrobial agents used as a first line and inexpensive treatment of BSI,such as ampicillin,penicillin,gentamicin,ciprofloxacin. The rise in antibiotic resistance in blood isolates emphasizes on rational and judicious use of antibiotics according to the antibiotic susceptibility/ resistance pattern of the institution. Specific antibiotic utilization strategies like antibiotic restriction, combination therapy and antibiotic recycling may help to decrease or prevent the emergence of resistance. Moreover there is need for strict aseptic precaution and sound infection control practices on the part of health care workers. These results also highlights the important role of local microbiology laboratories to detect resistance or reduced susceptibility in time to assist in evidence based antimicrobial treatment, to provide a baseline reference data for future studies, to detect changing trends in antimicrobial resistance pattern of pathogen at earliest thus can help in setting up priorities for focused intervention efforts and in formation of antibiotic prescribing and infection control policies of institution. Our result seem helpful in providing useful guidelines for choosing an effective antibiotic in cases of septicemia and for choosing salvage therapy against multidrug resistant strain.Our result should be interpreted cautiously since the study included a single referral hospital with few numbers of bacteria isolates as well as short study period.
ACKNOWLEDGEMENT
Authors acknowledge the immense help received from the scholars whose articles are cited and included in references of this manuscript. The authors are also grateful to authors / editors /publishers of all those articles, journals and books from where the literature for this article has been reviewed and discussed.
References:
1. Diekma DJ, Beekman SE, Chapin KC et al. Epidemiology and outcome of nosocomial and community onset bloodstream infection. j Clin Microbiol 2003; 41: 3655-60.
2. Young LS. Sepsis syndrome. In: Mandell GL, Bennett JE, Dolin R, eds. PrincipleandPracticeofinfectiousdiseases. Churchill Livingstone,1995; 690-705.
3. Warren DK, Zack JE, Elward AM, Cox MJ, Fraser VJ. Nosocomial primary bloodstream infections in intensive care unit patients in a non teaching community medical center : a 21-month prospective study. Clin Infect Dis 2001; 33 :1329-35.
4. Jang TN, Kuo BI, Shen SH, Fung CP, Lee SH, Yang TL, et al. Nosocomial Gram negative bacteremia in critically ill patients: epidemiologic characteristics and prognostic factors in 147episodes. Jformos Med Assoc 1999; 98 : 465-73
5. National Nosocomial Infections Surveillance (NNIS) system report,data summary from January 1992-April 2000, issued June 2000. AmJ Infect Control 2000; 28:429–48
6. Edmond MB, Wallace SE, McClish DK, Pfaller MA, Jones RN, Wenzel RP. Nosocomial bloodstream infections in United States hospitals: a three-year analysis. Clin Infect Dis 1999; 29:239–44
7. Diekema DJ, Pfaller MA, Jones RN. Agerelated trends in pathogen frequency and antimicrobial susceptibility of bloodstream isolates in North America. SENTRY Antimicrobial Surveillance Program,1997– 2000. Int J Antimicrob Agents 2002; 20:412-8.
8. Diekema DJ, Pfaller MA, Jones RN, Doern GV, Winokur PL, Gales AC, Sader HS, Kugler K, Beach M: Survey of bloodstream infections due to gram-negative bacilli: frequency of occurrence and antimicrobial susceptibility of isolates collected in the United States, Canada, and Latin America for the SENTRY antimicrobial surveillance program, 1997. Clin Infect Dis 1999; 29:595-607.
9. Garner JS, Jarvis WR, Emori TG, Horan TC, Hughes JM, CDC definations for nosocomial infections,1988.Am J Infect Control 1988;16:128-40..
10. National Committee for Clinical Laboratory Standards: Performance standards for antimicrobial susceptibility testing : 2009. In Supplemental tables, M100-S19. NCCLS, Wayne, PA; 2009
11. Ibrahim EH, Sherman G, Ward S, Fraser VJ, Kollef MH. The influence of inadequate antimicrobial treatment of bloodstream infections on patient outcomes in the ICU setting. Chest 2000; 118:146–55.
12. Falagas ME, Bakossi A, Pappas VD, et al. Secular trends of blood isolates in patients from rural area population hospitalized in tertiary center in a small city in Greece. BMC Microbiology 2006; 6: 41. Availabel at URL http://www.biomedcentral.com/1471- 2180/6/41
13. Roy I, Jain A , Kumar M, Agarwal SK:Bacteriology of Neonatal Septicaemia in a Tertiary Care Hospital of Northeren India. Indian Journal of Medical Microbiology, 2002;20:156-59.
14. Arora U Devi P. Bacterial profile of blood stream infections and antibiotic resistance pattern of isolates. J K Sci 2007; 9:186-190.
15. Garg A, Anupurba S, Garg J et al: Bacteriological Pro-file and Antimicrobial Resistance of Blood Culture Iso-lates from a University Hospital. JIACM 2007; 8 (2): 139-43.
16. Sharma M, Goel N, Chaudhary U Aggarwal R, Arora DR. Bacteraemia in children. Indian J Pediatr 2002; 69:1029-32
17. Kumhar GD, Ramachandran VG and Gupta P: Bacteriological Analysis of Blood Culture Isolates from Neonates in a Tertiary Care Hospital in India. J Health Popul Nutr 2002; 20 (4): 343-347.
18. Polymicrobial bacteraemias in England, Wales and Northern Ireland:2003. CDR Weekly 2003;14: No.51.
19. Nimra LF, Batchoun R. Communityacquired bacteraemia in a rural area: predominant bacterial species and antibiotic resistance. J Med Microbiology 2004; 53:1045-49.
20. Karlowsky JA, Jones ME, Draghi DC, Thornsberry C, Sahm DF, Volturo GA. Prevalence and antimicrobial susceptibilities of bacteria isolated from blood cultures of hospitalized patients in the United States in 2002. Annals of clinical microbiology and antimicrobials 2004;3:7.
21. Ahmed SH, Daef EA, Badary MS, Mahmoud MA, Abd-Elsayed AA.Nosocomial blood stream infection in intensive care units at Assiut University Hospitals (Upper Egypt) with special reference to extended spectrum β-lactamase producing organisms. BMC Research Notes 2009, 2:76
22. Wisplinghoff H, Bischoff T, Tallent SM, Seifert H, Wenzel RP, Edmond MB: Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin Infect Dis 2004, 39:309-17.
23. Narain S, Shastri JS, Mathur M, Mehta PR. Neonatal systemic candidiasis in a tertiary care centre. Indian Journal of Medical Microbiology, 2003;21:56-58
24. Chakrabarti A, Chander J , Kasturi P , Panigrahi D.Candidaemia: a 10 -year study in an Indian teaching hospital. Mycoses1992:35:47-51.
25. Kumar Surinder,Rizvi Meher,Vidhani Shalini,Sharma VK.Changing face of septicaemia and increasing drug resistance in blood isolates. Indian journal of medical microbiology,2003;21;56-58.
|






 This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License