IJCRR - 4(20), October, 2012
Pages: 39-49
Date of Publication: 20-Oct-2012
Print Article
Download XML Download PDF
DEGRADATION OF AZO AND TRIPHENYLMETHANE DYE BY THE BACTERIA ISOLATED FROM LOCAL INDUSTRIAL WASTE
Author: Pratiksha Pradhan, Harsh Dev Kumar, Gireesh Babu K
Category: General Sciences
Abstract:The local industrial sugar cane waste water sample was collected from Meerut, Uttar Pradesh for the isolation of dye degrading bacteria. The isolate SCWS5 was selected based on its maximum decolorization of dyes Methyl orange, Congo red and Malachite green (each 150mg/L). Phenotypic characterization and phylogenetic analysis based on 16S rDNA sequence confirmed SCWS5 as Bacillus sp. On physiochemical parameters optimization, Bacillus sp. SCWS5 degraded 10.13% (0.15mg/mL), 11.76% (0.11mg/mL) and 9.32% (0.8mg/mL) of Methyl orange, Congo red and Malachite green, respectively within 2h whilst 100% degradation (2.97mg/mL) at 18h for Methyl orange, 100% (2.43mg/mL) at 16h for Congo red and 100% (3.18mg/mL) at 20h for Malachite green were recorded. The degraded products were found non-toxic in nature while bacterial cells showed elongation and membrane rupturing under SEM taken from dye degraded media.
Keywords: Decolorization, Azo-dye, Triphenylmethane dye, SEM, Toxicity plasma parameters.
Full Text:
INTRODUCTION
Dyes are usually aromatic, heterocyclic compounds and often recalcitrant, some of them being toxic and even carcinogenic [1]. Various types of synthetic dyes and pigments are extensively used (approximately 100,000 tons/year) worldwide in textile sector [2]. During synthesis and usage, an estimated 10 to 15% of total dye content is released into the environment [3]. These are chemically diverse and divided into azo, anthraquinone, heterocyclic polymers and triphenylmethane dyes. Most of these are stable against light, temperature and biodegradation and have therefore accumulated in the environment as recalcitrant compounds [4]. Thus, the wastewater must be treated before releasing into the environment. For biological treatment of wastewater containing dyes, microbial decolorization and degradation of dyes has been of considerable interest [5]. Numerous studies have demonstrated on the ability of bacteria in monoculture to degrade dyes anaerobically and aerobically [6]. Many microorganisms are capable of degrading azo dyes, including bacteria, fungi and yeast [7]. Also the aerobic degradation of triphenylmethane dyes has been also demonstrated repeatedly; however these dyes resisted degradation in activated sludge systems [8]. Although research on biodegradation of different dyes by microbial consortia from polluted sites have been reported internationally but limited reports have been published on the dye degradation by the bacteria isolated from local industrial wastes. The present study emphasized on the degradation of three dyes belonging to monoazo, diazo and triphenylmethane group by the bacteria isolated from local industrial waste, which can be useful in providing an alternate method to accomplish dye degradation of a wide range of dyes in an eco-friendly manner.
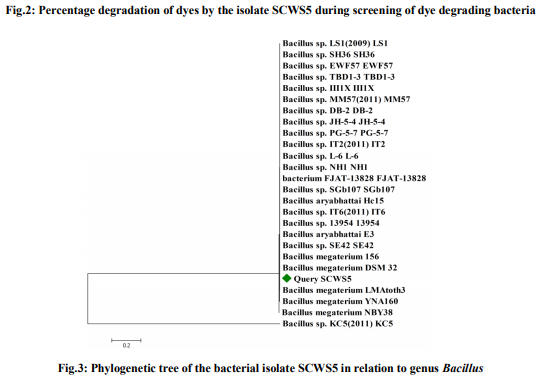
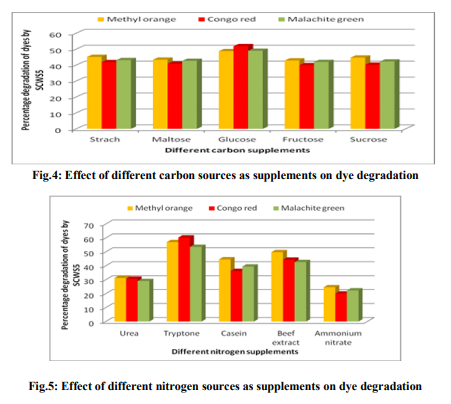
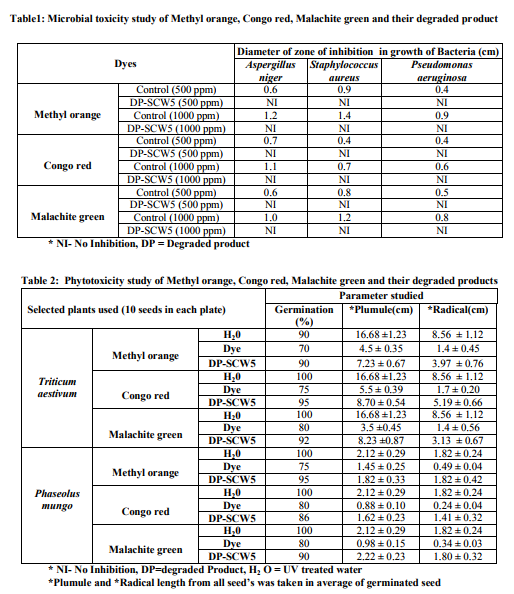
MATERIALS AND METHODS Dyes and chemicals The dyes Methyl orange (Fig.1a), Congo red (Fig.1b) and Malachite green (Fig.1c) used in the study were of analytical grade procured from Himedia, Mumbai, India. The Methyl orange is a mono azo dye of the 4- dimethylaminoazobenzene-4'-sulfonic acid sodium salt and Congo red is the sodium salt of benzidinediazo-bis-1-naphtylamine-4-sulfonic acid, a secondary diazo dye whereas Malachite green is the triphenylmethane dye of {4-[(4- dimethylaminophenyl)-phenyl-methylidene]- dimethyl-azanium chloride}. The Methyl orange, Congo red and Malachite green has maximum absorbance (λmax) of light at 444nm [9], 530nm [10] and 620nm [11], respectively. Isolation and identification of dye degrading bacteria The wastewater sample of local sugar cane industry collected from Meerut, Uttar Pradesh, India was screened for the dye degrading bacteria. The screening and routine sub culturing of dye decolorizing bacterial isolate was carried out on nutrient media (in g/L: Peptone, 10; Sodium chloride, 5; Yeast extract, 5) at pH 7 on 35ºC. The wastewater sample was tenfold serially diluted and 100µL of 10-5 dilution was spread plated on the nutrient agar plates. After 24h incubation, morphologically distinct colonies obtained were picked, purified by streak plate method and inoculated into the separate liquid medium amended with individual 150mg/L Methyl orange, Congo red and Malachite green dyes to analyze their decolorizing ability. Following 10h interval, aliquot of 1mL was withdrawn and centrifuged at 10,000rpm for 10min. The clear supernatant obtained was used to measure the dye decolorization at their respective absorbance maxima (λmax) using spectrophotometer where a medium without dye and inoculum was used as blank while medium with dye but without inoculum was taken as control. The absorbances for all the isolates were taken similarly up to 50h. All the experiments were carried out in triplicates and the mean value was taken. The decolorization of dyes was calculated by the given formula [12] as; Decolorization efficiency (%)=I-F/I ×100, Where I=Absorbance of media prior to incubation, F=Absorbance of decolorized media. After calculating the percentage decolorization of dyes up to 50h, the bacterial isolate with the strongest decolorizing ability, designated as SCWS5, was selected and preserved on slant and broth as stock. The isolate SCWS5 was initially studied for the phenotypic characteristics on the nutrient agar media followed by Gram staining [13] and then outsourced to Bioserve Biotechnologies Pvt. Ltd, Hyderabad, India, for molecular identification by 16S rDNA technique. The 16S rDNA sequence obtained was initially analyzed at NCBI server (http://www.ncbi.nlm.nih.gov/) using BLAST (Blastn) tool and corresponding sequences downloaded were then further used for phylogenetic analyses using MEGA version 4 [14]. Study of physiochemical parameters The isolated strain SCWS5 was further studied for the physiochemical parameters to check their effect on the dyes decolorization taking different carbon sources (1%), nitrogen sources (1%), pH (4, 5, 6, 7, 8, 9 and 10), temperature (25, 30, 35 and 40ºC), various dye concentrations (150, 200, 250, 300 and 350mg/L), different volumes of inoculum (100, 150, 200, 250 and 300µL) and static/shaker condition. The dyes decolorization was monitored as described in the earlier section. The decolorization analysis of the dyes was then further observed using all the optimized parameters with respect to time after every 2h interval while the cell pellets obtained were dried at 85°C and weighed mass was then expressed in mg/mL. Study of decolorization mode of SCWS5 on dyes and Scanning Electron Microscope Decolorization of dyes may take place by adsorption [15] or degradation [16]. Dye adsorption can be also easily judged by an evidently colored cell pellet, whereas those retaining their original colors are accompanied by the occurrence of biodegradation [7]. The three sets of optimized nutrient broth (25mL) inoculated (1% v/v) with pure bacterial isolate SCWS5 were incubated overnight and before addition of dyes (individually150mg/L in separate flask), one flask from each set were autoclaved at 121°C for half an hour while the unautoclaved served as control. After 24h incubation, samples from the flasks were withdrawn and centrifuged at 10,000rpm for 10min. The supernatant was used for monitoring the dye decolorization as described earlier. To determine the effect of dyes on the isolate SCWS5 cell morphology, visualization of the 24h old cultures grown in normal and individual dye (150mg/L) amended optimized nutrient broth were performed under scanning electron microscope (SEM) [17]. Toxicity study Microbial toxicity and phytotoxicity test were performed in order to assess the toxicity of dyes and their degraded products formed. The degraded products of each 150mg/L Methyl orange, Congo red and Malachite green from the 50mL optimized nutrient media were extracted in equal volume of ethyl acetate, dried and dissolved in 5mL sterilized distilled water to make a final concentration of 500ppm and 1000ppm. These final concentrations were then used for the microbial toxicity studies carried out in relation to Staphylococcus aureus, Pseudomonas aeruginosa and Aspergillus niger while the respective concentrations of dyes were taken as control. The growth inhibition zones of microbes (diameter in cm) were recorded after 24h and 48h incubation at 35°C for the bacterial and fungal strains, respectively. The phytotoxicity study of Methyl orange, Congo red, Malachite green and their degraded products (50mL dye degraded media extracted in ethyl acetate, dried and final concentration of 100ppm in 5mL) was also carried out (at room temp) in relation to Triticum aestivum and Phaseolus mungo (10 seeds each) by watering separately 5mL sample of control Methyl orange, Congo red and Malachite green and their degraded product (100ppm per day). The control set was watered with distilled water simultaneously whilst germination (%), length of plumule (shoot) and radicle (root) was recorded after 10 days. RESULTS Screening and identification of dye degrading bacteria A bacterial isolate SCWS5 was able to degrade 65.17% of Methyl orange, 69.41% of Congo red and 53.23 % of Malachite green within 50h incubation in individual dye amended nutrient broth (Fig.2). The isolate SCWS5 colonies were white, glossy, round with regular margins and 1 to 3mm in diameter and was observed to be Gram-positive rods. Sequence analysis of 16S rDNA showed that the isolate SCWS5 had highest similarity of 100% with Bacillus megaterium DMZ 32 (X60629). Based on the phenotypic characteristics and phylogenetic analysis, isolate SCWS5 was identified as Bacillus sp. Phylogenetic relationship between the isolate SCWS5 with different members of the Bacillus sp. are illustrated in the Fig.3. The 16S rDNA sequence of Bacillus sp. SCWS5 have been deposited in Gene Bank with accession number JQ900757. Analysis of physiochemical parameters Effect of carbon and nitrogen sources The dye decolorization efficiency of the bacterial strain SCWS5 was monitored with 1% different carbon and nitrogen sources as supplement in the nutrient media. Results concluded that among the different carbon sources, glucose showed maximum decolorization of 48.7, 51.8 and 49.9% for the Methyl orange, Congo red and Malachite green, respectively (Fig.4). Whilst among the different nitrogen supplements used, beef extract showed maximum decolorization of 49.7, 44.3 and 42.6% for the Methyl orange, Congo red and Malachite green, respectively (Fig.5).
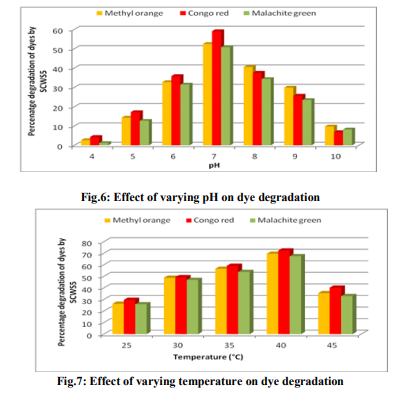
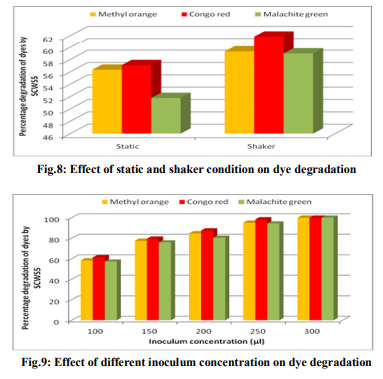
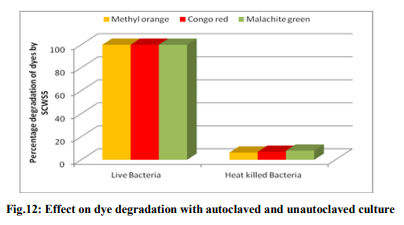
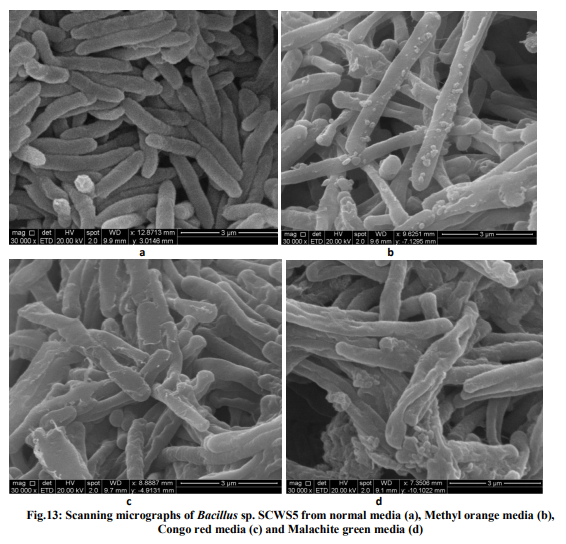
Effect of pH and temperature The strain SCWS5 represented lowest dye degradation of 2.67, 4.34 and 1.21% at pH 4 while maximum degradation of 52.4, 59 and 50.7% were observed for Methyl orange, Congo red and Malachite green, respectively at pH 7(Fig.6). The lowest dyes degradation of 26.2, 29.6 and 25.8% at temperature of 25°C while maximum dye degradation of 69.5, 72.3 and 67.4% at 40°C were observed for Methyl orange, Congo red and Malachite green, respectively (Fig.7). Decolorization on static and shaker conditions It was monitored that under static condition, isolate SCWS5 degraded 56.3, 57 and 51.7% of the Methyl orange, Congo red and Malachite green, respectively whereas higher degradation of 59.3, 61.8 and 58.9% for Methyl orange, Congo red and Malachite green, respectively were recorded on the shaker condition (Fig.8). Effect of inoculum and dye concentration The decolorization of 58.3, 61.3 and 57.1% were recorded for the 100µL volume of inoculum whereas 100% decolorization were recorded with the use of 300µL volume of inoculum for the 150 mg/L of Methyl orange, Congo red and Malachite green after 24h incubation at 35ºC (Fig.9). The study also recorded that at 150mg/L concentration the isolate showed 58.3, 61.3 and 57.1% decolorization which decreased to 14.2, 17.1 and 10.4% for the 350mg/L of Methyl orange, Congo red and Malachite green, respectively (Fig.10). Effect of total physiochemical parameters The decolorization efficiency of the bacterial isolate SCWS5 on individual 150mg/L dyes and its biomass was calculated using all of the optimized parameters with respect to time (Fig.11). It was deduced that bacteria degraded 10.1% (0.15mg/mL), 11.7% (0.11mg/mL) and 9.32% (0.08mg/mL) of Methyl orange, Congo red and Malachite green, respectively within 2h whilst 100% degradation (2.97mg/mL) at 18h for Methyl orange, 100% (2.43mg/mL) at 16h for Congo red and 100% (3.18mg/mL) at 20h for Malachite green were recorded. This proves that the combined effect of optimized parameters attributed to the decolourization of the dyes by the isolated bacteria Bacillus sp. SCWS5. Decolorization mode of Bacillus sp. SCWS5 and SEM analysis In the heat-killed bacterial cultures, only 6.22, 6.91and 7.87% decolorization of Methyl orange, Congo red and Malachite green were recorded after 24h incubation while in the control culture 100% decolorization of Methyl orange, Congo red and Malachite green were obtained in 24h (Fig.12) and the cell pellets were not pigmented. Moreover, the scanning micrographs showed elongation and cell membrane rupturing for cells taken from the dye degraded media as compared to the normal media as depicted in Fig.13 (a, b, c, d), revealed the toxic effect of dyes on the bacteria. Microbial toxicity and phytotoxicity study The inhibition zones were observed with control Methyl orange, Congo red and Malachite green with all bacterial strains studied, whereas their degraded products did not show any growth inhibition (Table1). During phytotoxicity study, germination (%) of both Triticum aestivum and Phaseolus mungo seeds were less in parent dyes compared to their respective degraded products, verified no toxicity of the degraded products (Table 2). DISCUSSION This study has been aimed at isolating and characterizing the potential dye degrading bacteria from the local industrial wastes. The strain Bacillus sp. SCWS5, isolated from sugarcane waste water used glucose and beef extract as carbon and nitrogen supplement respectively. Although effects of some other carbon sources on bacterial decolorization performance have been studied in former researches where lactate, peptone, succinate, yeast extract and formate were proved to enhance decolorization but sucrose and dextrin resulted in lower decolorization activities [18]. It was also reported that addition of inorganic nutrients like nitrogen does not always enhance degradation of organic compounds, because there are many other factors which many decrease microbial activity [19]. Urea when dissolved in liquid culture causes a shift of pH more towards acidic side, which decreased the color removal, growth as well as enzyme activity of strains. Also presence of nitrate in culture media might slow down color removal process [20]. The strain Bacillus sp. SCWS5 showed maximum dye decolorization at neutral pH, was in accordance with the report that optimum pH for color removal often exists between 6.0 and 10.0 [21]. Further, the isolate Bacillus sp. SCWS5 showed highest dye degradation on shaker must be because of dissolved oxygen concentration which creates the micro-aerophillic environment in the glass vessel for the bacteria bringing about the decolorization of dyes at a faster rate [22]. Results also concluded that for 100µL inoculum, 350mg/L dye acted as inhibitor for the bacterial growth and resulted in less decolorization whereas 300µL inoculum showed 100% decolorization of 150mg/L of dyes within 24h, suggested that the concentration of dye as substrate can influence the efficiency of dye removal through a combination of factors including the toxicity of dyes at higher concentrations and the ability of the enzyme to recognize the substrate efficiently at very low concentrations [23]. Further, the toxicity results for Methyl orange, Congo red and Malachite green coincides with the other research work reported for the respective dyes degradation using other bacteria [24, 10, 11]. The non-toxicity of dye degraded products can be suggested because of hydroxylase and oxygenase produced by the bacteria [25]. CONCLUSION It was inferred that the bacterial strain Bacillus sp. SCWS5 shows 100% degradation with the optimized physiochemical parameters. Since, the degraded products of the dyes showed no toxic effect on the microbes and plants, implies that maximum degradation of a wide range of different dyes can be meted out by employing the potential bacterial strains from the local industrial wastes using optimized physiochemical parameters as it renders the ability to bio transform the toxic dyes into non-toxic products without any additional treatment. This study further recommends the identification, purification of enzymes and their kinetics involved in the degradation of dyes by the isolate and exploitation of potential bacterial consortium from the local industrial wastes in the treatment of dye polluted waste water which would be cost-effective and can contribute to eradication of the dye pollution from the water bodies. ACKNOWLEDGEMENTS Ms. Pratiksha Pradhan one of the authors is thankful to Dr. J.J.D Pradhan, Director, Health and Family Welfare, Sikkim, for necessary support and assistance.
References:
1. Vyas BRM, Mollitoris HP. Involvement of an extracellular H2O2-dependent ligninolytic activity of the white rot fungus Pleurotus ostreatus in the decolorization of Remazol Brilliant Blue R. Appl Environ Microbiol 1995; 61:3919-3927.
2. Moreira MT, Mielgo I, Feijoo G, Lema JM. Evaluation of different fungal strains in the decolourization of synthetic dyes. Biotechnol Lett 2000; 22:1499-1503.
3. Verma P, Madamwar D. Comparative study of transformation of azo dye by different white rot fungi. Indian Journal of Biotechnology 2002; 1:393-396.
4. Vasdev K. Decolorization of triphenylmethane dyes by six white-rot fungi isolated from nature. Journal of Bioremediation and Biodegradation 2011; 29(5):128-133.
5. Kalme SD, Parshetti GK, Jadhav SU, Govindwar SP. Biodegradation of benzidine based dye Direct Blue 6 by Pseudomonas desmolyticum NCIM 2112. Bioresour Technol, doi:10.1016/j.biortech.2006.05.023.
6. Banat IM, Nigam P, Singh D, Marchant R. Microbial decolorization of textile dye containing effluents: A review. Bioresour Technol 1996; 58:217-227.
7. Chen KC, Wu JY, Liou DJ, Sz-Chwun JH. Decolorization of the textile dyes by newly isolated bacterial strains, J Biotechnol 2003; 10:57-68.
8. Sarnaik S, Kanekar P. Biodegradation of methyl Violet by Pseudomonas mendocina MCH B-402. Appl Environ Microbiol 1999; 52:251-254.
9. Chen Z-X, Jin X-Y, Chen Z, Megharaj M, Naidu R. Removal of methyl orange from aqueous solution using bentonite-supported nanoscale zero-valent iron. Journal of Colloid and Interface Science 2011; 363: 601-607.
10. Aileen C. Jalandoni B, Anna LA. Decena S, Ernelea PC, Virginia LB, Wilfredo LB. Characterization and identification of Congo red decolorizing bacteria from monocultures and consortia. Philippine Journal of Science 2010; 139:71-78.
11. Parshetti G, Kalme S, Saratale G, Govindwar S. Biodegradation of malachite green by Kocuria rosea MTCC 1532. Acta Chimica Slovenica 2006; 53(4):492-498.
12. Ola IO, Akintokun AK, Akpan I, Omomowo IO, Areo VO. Aerobic decolourization of two reactive azo dyes under varying carbon and nitrogen source by Bacillus cereus. Afr J Biotechnol 2010; 9(5):672-677.
13. Aneja KR. Experiments in Microbiology, Plant Pathology and Biotechnology, 4th. Ed. (2003). New Age International Publishers, New Delhi, India.
14. Tamura K, Dudley J, Nei M. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 2007; 24:1596-1599.
15. Aravindhan R, Rao JR, Nair BU. Removal of basic yellow dye from aqueous solution by sorption on green alga Caulerpa scalpelliformis. J Hazard Mater 2007; 142:68- 76.
16. Kumar K, Devi SS, Krishnamurthi K, Dutta D, Chakrabartiet T. Decolorisation and detoxification of Direct Blue-15 by a bacterial consortium. Bioresour Technol 2007; 98:3168-3171.
17. Kassongo J, Togo C A. The potential of whey in driving microbial fuel cells: A dual prospect of energy recovery and remediation. Afr J Biotechnol 2010; 9(46):7885-7890.
18. Xu M, Guo J, Zeng G, Zhong X, Sunet G. Decolorization of anthraquinone dye by Shewanella decolorationis S12. Appl Microbiol Biotechnol 2006; 71:246-251.
19. Steffensen SW, Alexander M. Role of competition for inorganic nutrients in biodegradation of mixtures of substrates, Appl Environ Microbiol 1995; 61: 2859–2862.
20. Carliell CM, Barday SJ, Nadidoo N, Buckley CA, Muuholland DA, Senior E. Microbial decolorization of a reactive azo dye under anaerobic conditions. Water SA (Pretoria) 1995; 21:61-69.
21. Guo JB, Zhou JT, Wang D, Tiana C, Wanga P, Uddina MS, Yua H. Biocalalyst effects of immobilized anthraquinone on the anaerobic reduction of azo dyes by the salt-tolerant bacteria. Water Res 2007; 41:426-432.
22. Hu TL. Degradation of azo dye RP2B by Pseudomonas Luteola. Water Sci Technol 1998; 38:299-306.
23. Pearce CI, Lloyd JR, Guthrie JT. The removal of colour from textile wastewater using whole bacterial cells: a review. Dyes and Pigments 2003; 58:179–196.
24. Parshetti GK, Telke AA, Kalyani DC, Govindwar S P. Decolorization and detoxification of sulfonated azo dye methyl orange by Kocuria rosea MTCC 1532. J Hazard Mater 2009; 176: 503-09.
25. Pandey A, Singh P, Iyengar L. Bacterial decolorization and degradation of azo dyes. International Biodeterioration and Biodegradation 2007; 59:73-84.


|






 This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License