IJCRR - Vol 09 issue 10 current issue , May, 2017
Pages: 44-48
Date of Publication: 27-May-2017
Print Article
Download XML Download PDF
Detection of Carbapenem-Resistant Klebsiella Pneumoniae Isolates from Clinical Specimens in Nnamdi Azikiwe University Teaching Hospital, Nnewi
Author: Onukwube Cyril Chinedu, Agbakoba Nneka Regina, Egwuatu Chukwudi Charles, Aghanya Iloduba Nnaemeka
Category: Healthcare
Abstract:Background: Carbapenem-resistant Klebsiella pneumoniae is emerging rapidly worldwide. The accurate and prompt identification of Klebsiella pneumoniae carbapenemase (KPC) is a key to limiting the spread of these bacteria. Increasingly, Klebsiella bacteria have developed antibiotic resistance most recently to the class of antibiotics known as carbapenems.
Aim: The study was done to detect carbapenem-resistant Klebsiella pneumonia isolates from clinical specimens in Nnamdi Azikiwe University Teaching Hospital, Nnewi, Nigeria.
Methodology: K. pneumoniae clinical isolates were collected and processed at Nnamdi Azikiwe University Teaching Hospital Medical Microbiology and Parasitology laboratory from August 2012 to February 2013. Fifty-two Klebsiella pneumoniae recovered from different specimens namely; wound swabs (14), urine (26), sputum (7), and endocervical swab/high vaginal swab (5) were analyzed. Antimicrobial susceptibility testing (AST) was determined by agar disc diffusion method using Mueller-Hinton agar according to Clinical and Laboratory Standards Institute recommendations. Modified Hodge test (MHT) was carried out on clinical isolates that showed reduced disc sizes to carbapenem and resistant to the third generation cephalosporin. Polymerase chain reaction (PCR) was carried out to detect the presence of blaKPC gene. Results: Antibiotics susceptibility results revealed that meropenem and Imipenem showed the highest sensitivity (100%) against the isolates tested. Ertapenem showed 94.2% sensitivity. K. pneumoniae was isolated more from female patients 30(57.7%) than from male patients 22(42.3%). The result of the single PCR also revealed the absence of blaKPC gene. The PCR and MHT results showed strong consistency when compared.
Conclusion: Care must be taken in the prescription and administration of carbapenem as it is the last resort for many bacterial infection. Infection control policies should be strictly adhered to.
Keywords: K. Pneumonia, Antimicrobial susceptibility testing (AST), Parasitology, Carbapenem
Full Text:
INTRODUCTION
Klebsiella pneumoniae is a Gram-negative, non-motile, encapsulated, lactose fermenting, facultative anaerobic, rod shaped bacterium found in the normal flora of the mouth, skin, and intestines (Ryan and Ray, 2004).
K. pneumoniae is an important cause of human infections or diseases which are usually nosocomial or hospital-acquired. Diseases caused by K. pneumoniae include; urinary tract infections, pneumonia, septicemia, and soft tissue infections (Podschun and Ullmann, 1998). The diseases caused by K. pneumoniae can result in death for patients who are immunodeficient. Capsular polysaccharide and lipopolysaccharide O side chain are two of the most important virulence factors of K. pneumoniae (Cortés et al., 2002).
Increasingly, Klebsiella bacteria have developed antibiotic resistance most recently to the class of antibiotics known as carbapenems (LA County Department of Public Health, 2010). Detection of carbapenemase-producing isolates in the microbiology laboratory is difficult (Tenover et al., 2006). Acquisition of a blaKPC gene alone does not always confer resistance to carbapenems, as defined by current breakpoints of the Clinical and Laboratory Standards Institute (CLSI). Some KPC isolates show low-level resistance to carbapenems; with elevated minimal inhibitory concentrations (MICs) (2 to 4 µg/mL) or with reduced disk zone diameters but within the susceptible range.
Emergence of carbapenem resistance in health care settings is increasingly attributed to the production of β-lactamases capable of hydrolysing carbapenems (Bilavsky et al., 2010). Carbapenemases are sub-class of β-lactamase enzymes that are classified by their specific resistant mechanisms, (such as AmpC and others), this research will concentrate on Klebsiella pneumoniae carbapenemase (KPC). The KPC enzyme confers resistance to all β-lactam agents including penicillins, cephalosporins, monobactams, and carbapenems (Smith et al., 2003; Alba et al., 2005).
KPC represents an emerging bacterial resistance mechanism and is currently more prevalent in the north-eastern part of the U.S. (New York, New Jersey and Maryland), although it has been seen with more frequency in other parts of the country (Martin et al., 2010). KPC-producing bacteria have also caused outbreaks in Israel and recently have become an emerging public health concerns in several regions worldwide, such as China, Latin America and Greece (Nordmann et al., 2009). Though there have been increasingly, reported cases of carbapenem-resistant Klebsiella pneumoniae worldwide, there is limited data in Africa. We decided to carry out this research due to the high incidence of medical tourism by Nigerians in places where KPC has been reported. Therefore, this work was centred on detection of Klebsiella pneumoniae carbapenemase as it is the prevalent and most documented mechanism of resistance by carbapenem-resistant Klebsiella pneumoniae.
Given the limited therapeutic options available, the accurate detection of KPC-possessing Klebsiella pneumoniae is crucial in controlling its spread. The current guidelines for the phenotypic detection of KPC-producing organisms in US hospitals are based on reduced susceptibility to carbapenems, which has to be confirmed by the Modified Hodge Test (MHT) (CLSI, 2009).
MATERIALS AND METHODS
Isolates Collection and Identification
Out of 702 bacterial isolates examined from Nnamdi Azikiwe University Teaching Hospital Medical Microbiology and Parasitology laboratory Nnewi, 52 were isolated as presumptive K. pneumoniae. Isolates were collected from urine, sputum, wound swab, high vaginal swab and endocervical swab samples. The collected isolates were processed using standard routine Microbiological methods. Isolation was done by sub-culturing the presumptive K. pneumoniae on MacConkey and EMB (Eosin Methylene Blue) agars. The plates were incubated at 37oC for 24hours. Confirmation of K. pneumoniae was based on morphology, gram staining and standard biochemical tests.
Antibiotics susceptibility testing
Antibiotics susceptibility testing was determined using the Kirby-Bauer disk diffusion method on Mueller-Hinton agar plates as recommended by Clinical and Laboratory Standard Institute (CLSI, 2009).
The standard commercial disks used and their concentrations are as follows: Ciprofloxacin (5µg), Imipenem (10µg), Ceftazidime (30µg), Cefotaxime (30µg), (Abtek biological, UK), Ertapenem (10µg), Aztreonam (30µg), Piperacillin/Tazobactam (10µg), Gentamicin (10µg), and Cefepime (30µg), (Oxoid, UK), Meropenem (10µg) (Mast Diagnostics ltd, UK).
Three to five discrete colonies of K. pneumoniae isolates selected from an 18-24hour agar plate were touched with a sterile wire loop and suspended in 5ml of sterile saline in a bijou bottle. The suspension was mixed thoroughly to obtain a homogenous mixture. The turbidity of the suspension was then adjusted to match the 0.5 McFarland turbidity standard.
Each of the isolates was uniformly and aseptically inoculated into a well dried Mueller-Hinton agar, in plates by spread plate method, as follows; a sterile swab stick was dipped in the suspension, squeezed by the side of the bottle before streaking over the sensitivity plates. The appropriate antibiotic discs were placed aseptically on the agar using sterile forceps. Inoculated plates were incubated at 37°C for 18-24hours after which the diameters of the inhibition zones were measured and recorded (as susceptible, intermediate or resistant). All susceptibility tests were carried out in duplicates. Klebsiella pneumoniae ATCC 13883 was used as control strain.
Modified Hodge Test
This is a phenotypic test which could be used to determine if reduced susceptibility to carbapenems is mediated by a carbapenemase. It’s the most easily performed confirmatory test for KPCs.
Modified Hodge test (MHT) was performed first by streaking a susceptible Escherichia coli isolate on a Mueller-Hinton plate, after which a carbapenem disk (Meropenem) was placed on the centre. Isolates suspected of carbapenemase production then were streaked from the disk to the outer margin of the plate. This test was also repeated in duplicates. Quality control was performed using Escherichia coli ATCC 25922, Klebsiella pneumoniae ATCC BAA 1705.
Molecular Analysis
DNA Amplification
Polymerase Chain Reaction (using the PCR machine; PTC-200 BY MJ Research) was carried out to detect the presence of blaKPC gene. The primers used are as follows; blaKPC-F1: TCGCTAAACTCGAACAGG and blaKPC-R1: TTACTGCCCGTTGACGCCCAATCC. The PCR reaction system contained 0.1-1.0µM for each primer. Cycling parameters were; initial denaturation at 93oC for 3 minutes, followed by 35 cycles of 1 minute at 93oC, 30 seconds at 56oC, and 1 minute 30 seconds at 72oC. The PCR amplification was ended by a final extension at 72oC for 10 minutes.
RESULTS
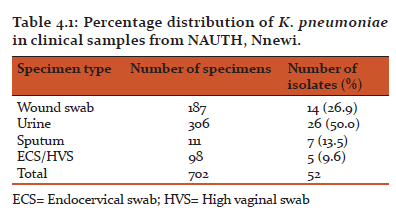
Of the 702 bacterial isolates analyzed for the presence of K. pneumoniae, 52 were identified as K. pneumoniae. Sources of the isolated K. pneumoniae were; wound swab 14(26.9%), urine 26(50.0%), sputum 7(13.5%), and ECS/HVS 5(9.6%), as shown in Table 4.1. The prevalence of K. pneumoniae clinical isolates was found to be 7.4%.
Table 4.1 shows the percentage distribution of K. pneumoniae in clinical samples from NAUTH.
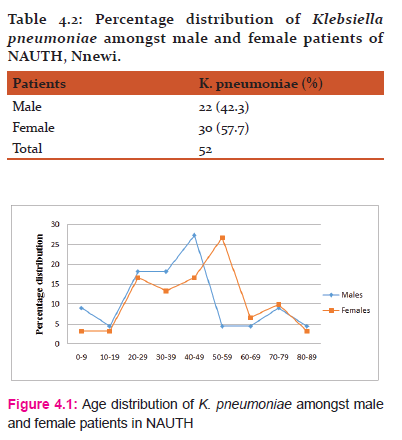
Table 4.2 outlined the percentage distribution of K. pneumoniae amongst male and female patients.
Fig. 4.1 shows the age distribution of K. pneumoniae amongst male and female patients in NAUTH. Among the male patients, age interval of 40-49 was the modal class, while the mean was 39.95. The modal class for the female patients was 50-59 while the mean was 45.83.
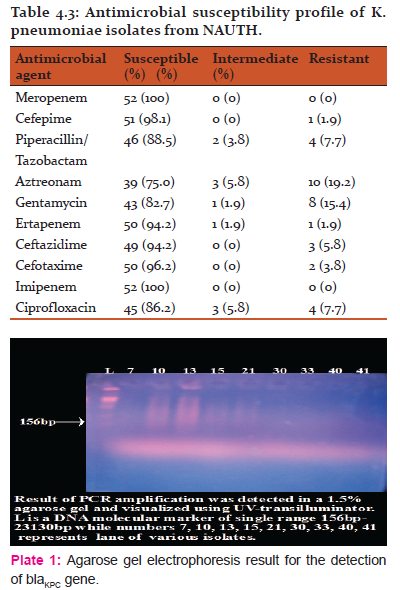
The Antimicrobial susceptibility pattern of the 52 Klebsiella pneumoniae clinical isolates was evaluated. Table 4.3 shows the results of the test.
10(19.2%) clinical isolates of K. pneumoniae that showed reduced susceptibility and resistance to third generation cephalosporin were subjected to polymerase chain reaction (PCR) to detect if blaKPC gene was present. Plate 1: shows the result. BlaKPC gene was not detected from the K. pneumoniae isolates tested.
DISCUSSION
With the increased dependence on carbapenem antibiotics, KPCs are a major public health concern. Bacterial isolates producing KPCs are able to hydrolyze a broad spectrum of β-lactams including the penicillins, cephalosporins, carbapenems and monobactams. They have the potentials to spread rapidly in hospital environments to cause nosocomial infections with high mortality rates (Hirsch and Tam, 2010). Carbapenems are a class of β-lactam antibiotics with a broad spectrum of antibacterial activity. They are one of the antibiotics of last resort for many bacterial infections, such as Escherichia coli (E. coli) and Klebsiella pneumoniae (Smith, 2010).
In this study, a total of 52 clinical isolates of Klebsiella pneumoniae were isolated from 702 clinical bacterial isolates. Sources of K. pneumoniae clinical isolates were as follows; wound swab, urine, sputum, and ECS/HVS. K. pneumoniae was predominantly isolated from urine samples 26(50%) followed by wound swab 14(26.9%), then sputum and ECS/HVS 7(13.5%) and 5(9.6%) respectively. This result conforms to the research done by Jombo et al., (2011) who reported urine specimen to have the highest number of Klebsiella isolates followed by wound swab or pus, sputum and then ECS/HVS. Iroha et al., (2011), reported that out of 390 samples collected, K. pneumoniae was isolated from 150 samples (urine 72, HVS 12, wound swab 16, and sputum 50). In their research, urine specimen constituted the largest number of K. pneumoniae isolates. Sarathbabu et al., (2012), also isolated K. pneumoniae predominantly from urine samples in hospitalised patients in India. K. pneumoniae is an opportunistic bacterium which affects the elderly and the immunocompromised. Therefore, the isolation of K. pneumoniae more frequently from urine specimen could be due to the presence of indwelling devices such as catheter in patients associated with the diseases. Presence of these devices in the body leads to the adherence of Gram-negative organisms such as Klebsiella on the surfaces of these devices which eventually promotes the establishment and development of the disease. Unlike the mucous membrane which serves as the first line of defence against bacteria and other pathogens, these invasive devices do not have such protective properties rather, it predisposes one to infections.
Antimicrobial susceptibility testing revealed that all K. pneumoniae isolates were susceptible to meropenem and Imipenem (100% sensitivity each). Fifty (94.2%) K. pneumoniae isolates showed susceptibility to ertapenem. High antimicrobial drug susceptibility pattern was also observed among other antimicrobial drugs used; Cefepime 51(98.1%), Piperacillin/tazobactam 46(88.5%), Aztreonam 39(75%), Gentamycin 43(82.75), Ceftazidime 49(94.2%), Cefotaxime 50(96.2%), and Ciprofloxacin 45(86.2%). This result is similar to that reported by Iroha et al., (2011), who in their research reported resistance to the commonly used antimicrobial agents such as chloramphenicol and ampicillin while carbapenems, aminoglycosides, cephalosporins, fluoroquinolones and monobactam showed greater sensitivity. This result is an indication that the antimicrobial drugs used in this study have not been grossly misused in this environment.
Varying degree of resistance (1-3 of the antimicrobial drugs used) was observed among the K. pneumoniae clinical isolates. Aztreonam showed highest resistance (19.2%) while lowest antimicrobial drug resistance was observed in cefepime (1.9%) and ertapenem (1.9%). It is worthy of note here that ertapenem non-susceptibility is not specific for carbapenemase production especially in areas where carbapenemase production is uncommon (Woodford et al., 2007). Among the carbapenems, K. pneumoniae showed no resistance to meropenem and imipenem. However, (1.9%) resistance and moderate susceptibility (1.9%) was observed in ertapenem. Meropenem and ertapenem is a more sensitive indicator than imipenem for the detection of KPC (CLSI) using disc diffusion.
Results from this research showed that K. pneumoniae infection was observed more in female patients (57.7%) than in their male counterpart (42.3%). The age distribution amongst the male patients showed that the age interval of 40-49 occurred more frequently (27.3%) and therefore, is the modal class while the mean was calculated to be 39.95. Amongst the female patients, the modal class was observed to be 50-59 while the calculated mean was 45.83. Age distribution pattern of subjects infected with K. pneumoniae ranged from 7months to 87years. There was no statistical significant difference between the distribution of K. pneumoniae isolates amongst male and female patients.
The outcome of the single PCR amplification showed that Klebsiella pneumoniae carbapenemase was not detected among Klebsiella pneumoniae isolates in NAUTH. Comparing the Modified Hodge Test (MHT) result and that of PCR, one can observe that both results showed 100% sensitivity for the detection of KPC activity. This finding is consistent with those of Lijun et al., (2012). Here Lijun and associates investigated the presence of K. pneumoniae carbapenemase genes from 159 clinical Gram-negative isolates resistant to several classes of β-lactam antibiotics. The sensitivity and specificity of MHT by this result have shown to exceed 90%; however, several reports have noted the occurrence of false positive results when the MHT was used to detect carbapenemase in ESBL-producing isolates (Carvalhaes et al., 2010; Wang et al., 2011). The absence of carbapenem-resistant Klebsiella pneumoniae (CRKP) in NAUTH Medical Microbiology and Parasitology Laboratory as of the time of this research could be as a result of the exorbitant prices of carbapenems which makes them not to be readily availability, and therefore not easily misused.
CONCLUSION
Based on the outcome of this study, Klebsiella pneumoniae demonstrated a high resistance to Aztreonam and Gentamycin compared to other antimicrobial drugs used. Therefore, care must be taken in the prescription and administration of these antimicrobial agents. Strict infection control policies (such as hand washing hygiene, use of gowns, contact precautions etc) should be put in place to control the growing incidence of antibiotics resistance. Antimicrobial drugs should be bought only from certified local manufacturers and importers. This will help to reduce the high level of counterfeit drugs in the country. Proper education of the public through jingles and adverts should also be considered to discourage self-medication. With the outcome of the antimicrobial susceptibility testing (AST), one can conclude that if these antimicrobial agents are used judiciously, it will go a long way in providing the desired treatment efficiencies and at the same time minimising resistance.
AKNOWLEDGEMENT
We sincerely acknowledge the technical assistance of the laboratory scientists and resident doctors in the Department of Medical Microbiology and Parasitology, Nnamdi Azikiwe University Teaching Hospital, Nnewi Campus, Nigeria. We also acknowledge all the scholars whose articles are cited and included in the references of this manuscript. We are equally grateful to authors / editors / publishers of all those articles, journals and books from where the literature for this article has been reviewed and discussed.
Source of funding: None
Conflict of interest: Nil
References:
Alba, J., Ishii, Y., Thomson, K., Moland, E.S., and Yamaguchi, K. (2005). Kinetics study of KPC-3, a plasmid-encoded class A carbapenem-hydrolyzing β-lactamase. Antimicrobial Agents and Chemotherapy; 49: 4760-4762.
Bilavsky, E., Schwaber, M.J., and Carmeli, Y. (2010). How to stem the tide of carbapenemase-producing Enterobacteriaceae?: proactive versus reactive strategies. Current Opinion in Infectious Diseases; 23: 327-331.
Carvalhaes, C.G., Picao, R.C., Nicoletti, A.G., Xavier, D.E., and Gales, A.C. (2010). Cloverleaf test (modified Hodge test) for detecting carbapenemase production in Klebsiella pneumoniae: be aware of false positive results. Journal of Antimicrobial Chemotherapy; 65: 249-251
Clinical and Laboratory Standards Institute (2009). Performance standards for antimicrobial susceptibility testing; Nineteenth Informational Supplement. CLSI Document M100- S19. Wayne, PA: Clinical and Laboratory Standards Institute.
Cortés, G., Borrell, N., De-astorza, B., Gómez, C., Sauleda, J., and Albertí, S. (2002). "Molecular Analysis of the Contribution of the Capsular Polysaccharide and the Lipopolysaccharide O Side Chain to the Virulence of Klebsiella pneumoniae in a Murine Model of Pneumonia". Infection and Immunity; 70(5): 2583-2590.
Hirsch, B., and Tam, V.H. (2010). Detection and treatment options for Klebsiella pneumoniae carbapenemases (KPCs): an emerging cause of multidrug infection. Journal of antimicrobial chemotherapy; 65: 1119-1125.
Iroha, I.R., Oji, A.E., and Ayogu, T.E. (2011). Analysis of Antibiotic susceptibility of Klebsiella pneumoniae strains isolated from different clinical specimens in Enugu state. International Journal of Current Research; 2(1): 8-14.
Jumbo, G.T.A., Emanghe, U.E., Akpan, S., and Bolarin, D.M. (2011). Klebsiella infections and Antimicrobial susceptibility patterns in the 21st century: A Paradigm shift. Asian journal of Pharmaceutical and Health Sciences; 1(4): 204-208.
LA county Department of Public Health (2010). Acute communicable disease control: carbapenem-resistant Klebsiella pneumoniae. Retrieved November 15, 2012 from http://www.publichealth.lacounty.gov/acd/Diseases/Klebsiella.htm
Lijun, W., Haitong, G., and Xinxin, L. (2012). A Rapid Low-cost Real-time PCR for the Detection of Klebsiella pneumoniae Carbapenemase Genes. Annals of Clinical Microbiology and Antimicrobials; 11(9): 1-6.
Martin, G.J., and Friedlander, A.M. (2010). Anthrax as an agent of bioterrorism. In: Mandell G.L., Bennett, J.E. and Dolin, R. editors. Mandell, Douglas, and Bennetts’s Principles and Practice of Infectious Diseases. 7th ed. Philadelphia: Churchill Livingstone Elsevier. 3983-3992.
Nordmann, P., Cuzon, G., and Naas, T. (2009). The real threat of Klebsiella pneumoniae carbapenemase-producing bacteria. Lancet Infection Disease; 9: 228–236.
Podschun, R. and Ullmann, U. (1998). "Klebsiella spp. as Nosocomial Pathogens: Epidemiology, Taxonomy, Typing Methods, and Pathogenicity Factors". Clinical Microbiology Reviews; 11(4): 589-603.
Ryan, K.J. and Ray, C.G. (2004). Sherris Medical Microbiology (4th ed.). McGraw Hill.
Sarathbabu, R., Ramani, T.V., Bhaskara, R.K., and Supriya, P. (2012). Antibiotic susceptibility pattern of Klebsiella pneumoniae isolated from sputum, urine and pus samples. Journal of Pharmacy and Biological Sciences; 1(2): 4-9.
Smith, M., Hanson, E.N.D., Herrera, V.L., Black, J.A., Lockhart, T. J., Hossain, A., Johnson, J. A., Goering, R.V., and Thomson, K. S. (2003). Plasmid-mediated, carbapenem-hydrolysing β-lactamase, KPC-2, in Klebsiella pneumoniae isolates. Journal of Antimicrobial Chemotherapy; 51: 711-714.
Smith, S. (2010). "Deadly bacteria's foothold spurs study: Mass. specialists try to halt spread". Retrieved December 5, 2012, from http://www.boston.com/news/health/articles/2010/10/07/mass_specialists_to_study drug_resistant_germs_spread/
Tenover, F.C., Kalsi, R.K., and Williams, P.P. (2006). Carbapenem resistance in Klebsiella pneumoniae not detected by automated susceptibility testing. Emerging Infectious Diseases; 12: 1209-1213.
Wang, P., Chen, S., Guo, Y., Xiong, Z., Hu, F., Zhu, D., and Zhang, Y. (2011). Occurrence of false positive results for the detection of carbapenemases in carbapenemase-negative Escherichia coli and Klebsiella pneumoniae isolates. 6: 26356.
Woodford, N., Dallow, J.W., Hill, R.L., Palepou, M. F., Pike, R., Ward, M.E., Warner, M., and Livermore, D.M. (2007). Ertapenem resistance among Klebsiella and Enterobacter submitted in the UK to a reference laboratory. International Journal of Antimicrobial Agents; 29: 456-459.
|






 This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License